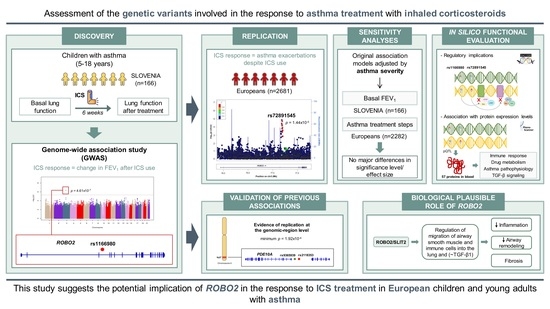
Identification of ROBO2 as a Potential Locus Associated with Inhaled Corticosteroid Response in Childhood Asthma
Abstract
:1. Introduction
2. Results
2.1. Characteristics of the Study Populations
2.2. Association Results of the Change in FEV1 after ICS Treatment
2.3. Validation of the Association with Asthma Exacerbations despite ICS Use
2.4. Sensitivity Analyses Accounting for Asthma Severity
2.5. In Silico Evaluation of the Variants Associated with Different Definitions of ICS Response
2.6. Validation of Previous Associations with ICS Response
3. Discussion
4. Materials and Methods
4.1. Study Population Analyzed in the Discovery Phase
4.2. Genotyping and Imputation of Genetic Variants in SLOVENIA
4.3. Association Testing with the Change in FEV1 Defined as a Binary Variable
4.4. Association with the Quantitative Change in FEV1 after ICS Treatment
4.5. Replication of Results Analyzing the Association with Asthma Exacerbations despite ICS Use in Additional Studies
4.6. Sensitivity Analyses of Asthma Treatment Response
4.7. In Silico Functional Evaluation of Variants Associated with ICS Response
4.8. Validation of Previous Associations with ICS Response
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Global Initiative for Asthma. Global Strategy for Asthma Management and Prevention. 2020. Available online: https://ginasthma.org/ (accessed on 25 October 2020).
- Cerasoli, F., Jr. Developing the Ideal Inhaled Corticosteroid. Chest 2006, 130 (Suppl. 1), 54S–64S. [Google Scholar]
- Yang, J.; Benyamin, B.; McEvoy, B.P.; Gordon, S.; Henders, A.K.; Nyholt, D.R.; Madden, P.A.; Heath, A.C.; Martin, N.G.; Montgomery, G.W.; et al. Common SNPs Explain a Large Proportion of the Heritability for Human Height. Nat. Genet. 2010, 42, 565–569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szefler, S.J.; Phillips, B.R.; Martinez, F.D.; Chinchilli, V.M.; Lemanske, R.F.; Strunk, R.C.; Zeiger, R.S.; Larsen, G.; Spahn, J.D.; Bacharier, L.B.; et al. Characterization of Within-Subject Responses to Fluticasone and Montelukast in Childhood Asthma. J. Allergy Clin. Immunol. 2005, 115, 233–242. [Google Scholar] [PubMed]
- Scelfo, C.; Galeone, C.; Bertolini, F.; Caminati, M.; Ruggiero, P.; Facciolongo, N.; Menzella, F. Towards Precision Medicine: The Application of Omics Technologies in Asthma Management. F1000Research 2018, 7, 423. [Google Scholar] [CrossRef] [PubMed]
- Dijkstra, A.; Vonk, J.M.; Jongepier, H.; Koppelman, G.H.; Koppelman, J.P.; ten Hacken, N.H.T.; Timens, W.; Postma, D.S. Lung Function Decline in Asthma: Association with Inhaled Corticosteroids, Smoking and Sex. Thorax 2006, 61, 105–110. [Google Scholar]
- Drazen, J.M.; Silverman, E.K.; Lee, T.H. Heterogeneity of Therapeutic Responses in Asthma. Br. Med. Bull. 2000, 56, 1054–1070. [Google Scholar]
- Mersha, T.B. Mapping Asthma-Associated Variants in Admixed Populations. Front. Genet. 2015, 6, 292. [Google Scholar]
- Ramadan, A.A.; Gaffin, J.M.; Israel, E.; Phipatanakul, W. Asthma and Corticosteroid Responses in Childhood and Adult Asthma. Clin. Chest Med. 2019, 40, 163–177. [Google Scholar]
- Fitzpatrick, A.M.; Teague, W.G.; Meyers, D.A.; Peters, S.P.; Li, X.; Li, H.; Wenzel, S.E.; Aujla, S.; Castro, M.; Bacharier, L.B.; et al. Heterogeneity of Severe Asthma in Childhood: Confirmation by Cluster Analysis of Children in the National Institutes of Health/National Heart, Lung, and Blood Institute Severe Asthma Research Program. J. Allergy Clin. Immunol. 2011, 127, 382–389.e13. [Google Scholar]
- Moore, W.C.; Hastie, A.T.; Li, X.; Li, H.; Busse, W.W.; Jarjour, N.N.; Wenzel, S.E.; Peters, S.P.; Meyers, D.A.; Bleecker, E.R. Sputum Neutrophil Counts Are Associated with More Severe Asthma Phenotypes Using Cluster Analysis. J. Allergy Clin. Immunol. 2014, 133, 1557–1563.e5. [Google Scholar] [CrossRef] [Green Version]
- Park, H.W.; Tantisira, K.G.; Weiss, S.T. Pharmacogenomics in Asthma Therapy: Where Are We and Where Do We Go? Annu. Rev. Pharmacol. Toxicol. 2015, 55, 129–147. [Google Scholar] [CrossRef]
- Duong-Thi-Ly, H.; Nguyen-Thi-Thu, H.; Nguyen-Hoang, L.; Nguyen-Thi-Bich, H.; Craig, T.J.; Duong-Quy, S. Effects of Genetic Factors to Inhaled Corticosteroid Response in Children with Asthma: A Literature Review. J. Int. Med. Res. 2017, 45, 1818–1830. [Google Scholar] [CrossRef] [Green Version]
- Al Moamary, M.S.; Al-Kordi, A.G.; Al Ghobain, M.O.; Tamim, H.M. Utilization and Responsiveness of the Asthma Control Test (ACT) at the Initiation of Therapy for Patients with Asthma: A Randomized Controlled Trial. BMC Pulm. Med. 2012, 12, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gorelick, M.H.; Stevens, M.W.; Schultz, T.R.; Scribano, P.V. Performance of a Novel Clinical Score, the Pediatric Asthma Severity Score (PASS), in the Evaluation of Acute Asthma. Acad. Emerg. Med. 2004, 11, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Keogh, K.A.; Macarthur, C.; Parkin, P.C.; Stephens, D.; Arseneault, R.; Tennis, O.; Bacal, L.; Schuh, S. Predictors of Hospitalization in Children with Acute Asthma. J. Pediatr. 2001, 139, 273–277. [Google Scholar] [CrossRef] [PubMed]
- Fuhlbrigge, A.; Peden, D.; Apter, A.J.; Boushey, H.A.; Camargo, C.A., Jr.; Gern, J.; Heymann, P.W.; Martinez, F.D.; Mauger, D.; Teague, W.G.; et al. Asthma Outcomes: Exacerbations. J. Allergy Clin. Immunol. 2012, 129 (Suppl. 3), S34–S48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- National Asthma Education and Prevention Program, Third Expert Panel on the Diagnosis and Management of Asthma. Expert Panel Report 3: Guidelines for the Diagnosis and Management of Asthma. Section 2, Definition, Pathophysiology and Pathogenesis of Asthma, and Natural History of Asthma; National Heart, Lung, and Blood Institute: Bethesda, MD, USA, 2007.
- Aldington, S.; Beasley, R. Asthma Exacerbations. 5: Assessment and Management of Severe Asthma in Adults in Hospital. Thorax 2007, 62, 447–458. [Google Scholar] [CrossRef] [Green Version]
- Gorelick, M.H.; Stevens, M.W.; Schultz, T.; Scribano, P.V. Difficulty in Obtaining Peak Expiratory Flow Measurements in Children with Acute Asthma. Pediatr. Emerg. Care 2004, 20, 22–26. [Google Scholar] [CrossRef]
- Szefler, S.J.; Martin, R.J.; King, T.S.; Boushey, H.A.; Cherniack, R.M.; Chinchilli, V.M.; Craig, T.J.; Dolovich, M.; Drazen, J.M.; Fagan, J.K.; et al. Significant Variability in Response to Inhaled Corticosteroids for Persistent Asthma. J. Allergy Clin. Immunol. 2002, 109, 410–418. [Google Scholar] [CrossRef] [Green Version]
- Martin, R.J.; Szefler, S.J.; King, T.S.; Kraft, M.; Boushey, H.A.; Chinchilli, V.M.; Craig, T.J.; Dimango, E.A.; Deykin, A.; Fahy, J.V.; et al. The Predicting Response to Inhaled Corticosteroid Efficacy (PRICE) Trial. J. Allergy Clin. Immunol. 2007, 119, 73–80. [Google Scholar] [CrossRef] [Green Version]
- Gallucci, M.; Carbonara, P.; Pacilli, A.M.G.; di Palmo, E.; Ricci, G.; Nava, S. Use of Symptoms Scores, Spirometry, and Other Pulmonary Function Testing for Asthma Monitoring. Front. Pediatr. 2019, 7, 54. [Google Scholar] [CrossRef] [Green Version]
- Cooper, B.G. Limitations to Spirometry Being Performed in “the Office”. Chron. Respir. Dis. 2005, 2, 113–115. [Google Scholar] [CrossRef] [Green Version]
- Tepper, R.S.; Wise, R.S.; Covar, R.; Irvin, C.G.; Kercsmar, C.M.; Kraft, M.; Liu, M.C.; O’Connor, G.T.; Peters, S.P.; Sorkness, R.; et al. Asthma Outcomes: Pulmonary Physiology. J. Allergy Clin. Immunol. 2012, 129 (Suppl. 3), S65–S87. [Google Scholar] [CrossRef] [Green Version]
- Hernandez-Pacheco, N.; Flores, C.; Oh, S.S.; Burchard, E.G.; Pino-Yanes, M. What Ancestry Can Tell Us About the Genetic Origins of Inter-Ethnic Differences in Asthma Expression. Curr. Allergy Asthma Rep. 2016, 16, 53. [Google Scholar] [CrossRef]
- Hernandez-Pacheco, N.; Pino-Yanes, M.; Flores, C. Genomic Predictors of Asthma Phenotypes and Treatment Response. Front. Pediatr. 2019, 7, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tantisira, K.G.; Damask, A.; Szefler, S.J.; Schuemann, B.; Markezich, A.; Su, J.; Klanderman, B.; Sylvia, J.; Wu, R.; Martinez, F.; et al. Genome-Wide Association Identifies the T Gene as a Novel Asthma Pharmacogenetic Locus. Am. J. Respir. Crit. Care Med. 2012, 185, 1286–1291. [Google Scholar] [CrossRef] [Green Version]
- Tantisira, K.G.; Lasky-Su, J.; Harada, M.; Murphy, A.; Litonjua, A.A.; Himes, B.E.; Lange, C.; Lazarus, R.; Sylvia, J.; Klanderman, B.; et al. Genome-wide Association between GLCCI1 and Response to Glucocorticoid Therapy in Asthma. N. Engl. J. Med. 2011, 365, 1173–1183. [Google Scholar] [CrossRef] [Green Version]
- Wu, A.C.; Himes, B.E.; Lasky-Su, J.; Litonjua, A.; Peters, S.P.; Lima, J.; Kubo, M.; Tamari, M.; Nakamura, Y.; Qiu, W.; et al. Inhaled Corticosteroid Treatment Modulates ZNF432 Gene Variant’s Effect on Bronchodilator Response in Asthmatics. J. Allergy Clin. Immunol. 2014, 133, 723–728.e3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, H.W.; Dahlin, A.; Tse, S.; Duan, Q.L.; Schuemann, B.; Martinez, F.D.; Peters, S.P.; Szefler, S.J.; Lima, J.J.; Kubo, M.; et al. Genetic Predictors Associated with Improvement of Asthma Symptoms in Response to Inhaled Corticosteroids. J. Allergy Clin. Immunol. 2014, 133, 664–669.e5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, T.J.; Park, J.S.; Cheong, H.S.; Park, B.L.; Kim, L.H.; Heo, J.S.; Kim, Y.K.; Kim, K.U.; Uh, S.T.; Lee, H.S.; et al. Genome-Wide Association Study Identifies ALLC Polymorphisms Correlated with FEV1 Change by Corticosteroid. Clin. Chim. Acta 2014, 436, 20–26. [Google Scholar] [CrossRef] [PubMed]
- Dahlin, A.; Denny, J.; Roden, D.M.; Brilliant, M.H.; Ingram, C.; Kitchner, T.E.; Linneman, J.G.; Shaffer, C.M.; Weeke, P.; Xu, H.; et al. CMTR1 Is Associated with Increased Asthma Exacerbations in Patients Taking Inhaled Corticosteroids. Immun. Inflamm. Dis. 2015, 3, 350–359. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tong, C.; Wang, Z.; Mauger, D.; Tantisira, K.G.; Israel, E.; Szefler, S.J.; Chinchilli, V.M.; Boushey, H.A.; Lazarus, S.C.; et al. Pharmacodynamic Genome-Wide Association Study Identifies New Responsive Loci for Glucocorticoid Intervention in Asthma. Pharmacogenom. J. 2015, 15, 422–429. [Google Scholar] [CrossRef] [Green Version]
- Leusink, M.; Vijverberg, S.J.; Koenderman, L.; Raaijmakers, J.A.; de Jongste, J.C.; Sterk, P.J.; Duiverman, E.J.; Onland-Moret, N.C.; Postma, D.S.; de Boer, A.; et al. Genetic Variation in Uncontrolled Childhood Asthma despite ICS Treatment. Pharmacogenom. J. 2016, 16, 158–163. [Google Scholar] [CrossRef] [PubMed]
- Mosteller, M.; Hosking, L.; Murphy, K.; Shen, J.; Song, K.; Nelson, M.; Ghosh, S. No Evidence of Large Genetic Effects on Steroid Response in Asthma Patients. J. Allergy Clin. Immunol. 2017, 139, 797–803.e7. [Google Scholar] [CrossRef] [PubMed]
- Levin, A.M.; Gui, H.; Hernandez-Pacheco, N.; Yang, M.; Xiao, S.; Yang, J.J.; Hochstadt, S.; Barczak, A.J.; Eckalbar, W.L.; Rynkowski, D.; et al. Integrative Approach Identifies Corticosteroid Response Variant in Diverse Populations with Asthma. J. Allergy Clin. Immunol. 2019, 143, 1791–1802. [Google Scholar] [CrossRef]
- Hernandez-Pacheco, N.; Farzan, N.; Francis, B.; Karimi, L.; Repnik, K.; Vijverberg, S.J.; Soares, P.; Schieck, M.; Gorenjak, M.; Forno, E.; et al. Genome-Wide Association Study of Inhaled Corticosteroid Response in Admixed Children with Asthma. Clin. Exp. Allergy 2019, 49, 789–798. [Google Scholar] [CrossRef] [Green Version]
- Hernandez-Pacheco, N.; Vijverberg, S.J.; Herrera-Luis, E.; Li, J.; Sio, Y.Y.; Granell, R.; Corrales, A.; Maroteau, C.; Lethem, R.; Perez-Garcia, J.; et al. Genome-Wide Association Study of Asthma Exacerbations despite Inhaled Corticosteroids Use. Eur. Respir. J. 2020, 57, 2003388. [Google Scholar] [CrossRef]
- Garcia-Menaya, J.M.; Cordobes-Duran, C.; Garcia-Martin, E.; Agundez, J.A.G. Pharmacogenetic Factors Affecting Asthma Treatment Response. Potential Implications for Drug Therapy. Front. Pharmacol. 2019, 10, 520. [Google Scholar] [CrossRef]
- Berce, V.; Kozmus, C.E.; Potocnik, U. Association among ORMDL3 Gene Expression, 17q21 Polymorphism and Response to Treatment with Inhaled Corticosteroids in Children with Asthma. Pharmacogenom. J. 2013, 13, 523–529. [Google Scholar] [CrossRef]
- Hernandez-Pacheco, N.; Gorenjak, M.; Jurgec, S.; Corrales, A.; Jorgensen, A.; Karimi, L.; Vijverberg, S.J.; Berce, V.; Schieck, M.; Acosta-Herrera, M.; et al. Combined Analysis of Transcriptomic and Genetic Data for the Identification of Loci Involved in Glucocorticosteroid Response in Asthma. Allergy 2020, 76, 1238–1243. [Google Scholar] [CrossRef]
- Farzan, N.; Vijverberg, S.J.; Andiappan, A.K.; Arianto, L.; Berce, V.; Blanca-Lopez, N.; Bisgaard, H.; Bonnelykke, K.; Burchard, E.G.; Campo, P.; et al. Rationale and Design of the Multiethnic Pharmacogenomics in Childhood Asthma Consortium. Pharmacogenomics 2017, 18, 931–943. [Google Scholar] [CrossRef]
- British Thoracic Society and the Scottish Intercollegiate Guidelines Network. British guideline on the management of asthma. Thorax 2014, 69 (Suppl. 1), 1–192. [Google Scholar]
- The ENCODE Project Consortium. An Integrated Encyclopedia of DNA Elements in the Human Genome. Nature 2012, 489, 57–74. [Google Scholar] [CrossRef]
- Staley, J.R.; Blackshaw, J.; Kamat, M.A.; Ellis, S.; Surendran, P.; Sun, B.B.; Paul, D.S.; Freitag, D.; Burgess, S.; Danesh, J.; et al. PhenoScanner: A Database of Human Genotype-Phenotype Associations. Bioinformatics 2016, 32, 3207–3209. [Google Scholar] [CrossRef] [Green Version]
- Kamat, M.A.; Blackshaw, J.A.; Young, R.; Surendran, P.; Burgess, S.; Danesh, J.; Butterworth, A.S.; Staley, J.R. PhenoScanner V2: An Expanded Tool for Searching Human Genotype-Phenotype Associations. Bioinformatics 2019, 35, 4851–4853. [Google Scholar] [CrossRef] [Green Version]
- Suhre, K.; Arnold, M.; Bhagwat, A.M.; Cotton, R.J.; Engelke, R.; Raffler, J.; Sarwath, H.; Thareja, G.; Wahl, A.; DeLisle, R.K.; et al. Connecting Genetic Risk to Disease End Points through the Human Blood Plasma Proteome. Nat. Commun. 2017, 8, 14357. [Google Scholar] [CrossRef] [Green Version]
- Sun, B.B.; Maranville, J.C.; Peters, J.E.; Stacey, D.; Staley, J.R.; Blackshaw, J.; Burgess, S.; Jiang, T.; Paige, E.; Surendran, P.; et al. Genomic Atlas of the Human Plasma Proteome. Nature 2018, 558, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Folkersen, L.; Fauman, E.; Sabater-Lleal, M.; Strawbridge, R.J.; Frånberg, M.; Sennblad, B.; Baldassarre, D.; Veglia, F.; Humphries, S.E.; Rauramaa, R.; et al. Mapping of 79 Loci for 83 Plasma Protein Biomarkers in Cardiovascular Disease. PLoS Genet. 2017, 13, e1006706. [Google Scholar] [CrossRef] [PubMed]
- Tirado-Rodriguez, B.; Ortega, E.; Segura-Medina, P.; Huerta-Yepez, S. TGF- Beta: An Important Mediator of Allergic Disease and a Molecule with Dual Activity in Cancer Development. J. Immunol. Res. 2014, 2014, 318481. [Google Scholar] [CrossRef] [Green Version]
- Carvalho-Silva, D.; Pierleoni, A.; Pignatelli, M.; Ong, C.; Fumis, L.; Karamanis, N.; Carmona, M.; Faulconbridge, A.; Hercules, A.; McAuley, E.; et al. Open Targets Platform: New Developments and Updates Two Years On. Nucleic Acids Res. 2019, 47, D1056–D1065. [Google Scholar] [CrossRef] [PubMed]
- Li, H.S.; Chen, J.H.; Wu, W.; Fagaly, T.; Zhou, L.; Yuan, W.; Dupuis, S.; Jiang, Z.H.; Nash, W.; Gick, C.; et al. Vertebrate Slit, a Secreted Ligand for the Transmembrane Protein Roundabout, Is a Repellent for Olfactory Bulb Axons. Cell 1999, 96, 807–818. [Google Scholar] [CrossRef] [Green Version]
- Kidd, T.; Brose, K.; Mitchell, K.J.; Fetter, R.D.; Tessier-Lavigne, M.; Goodman, C.S.; Tear, G. Roundabout Controls Axon Crossing of the CNS Midline and Defines a Novel Subfamily of Evolutionarily Conserved Guidance Receptors. Cell 1998, 92, 205–215. [Google Scholar] [CrossRef] [Green Version]
- Xian, J.; Clark, K.J.; Fordham, R.; Pannell, R.; Rabbitts, T.H.; Rabbitts, P.H. Inadequate Lung Development and Bronchial Hyperplasia in Mice with a Targeted Deletion in the Dutt1/Robo1 Gene. Proc. Natl. Acad. Sci. USA 2001, 98, 15062–15066. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dickinson, R.E.; Duncan, W.C. The SLIT-ROBO Pathway: A Regulator of Cell Function with Implications for the Reproductive System. Reproduction 2010, 139, 697–704. [Google Scholar] [CrossRef] [Green Version]
- Tole, S.; Mukovozov, I.M.; Huang, Y.W.; Magalhaes, M.A.; Yan, M.; Crow, M.R.; Liu, G.Y.; Sun, C.X.; Durocher, Y.; Glogauer, M.; et al. The Axonal Repellent, Slit2, Inhibits Directional Migration of Circulating Neutrophils. J. Leukoc. Biol. 2009, 86, 1403–1415. [Google Scholar] [CrossRef] [Green Version]
- Lin, Y.Z.; Zhong, X.N.; Chen, X.; Liang, Y.; Zhang, H.; Zhu, D.L. Roundabout Signaling Pathway Involved in the Pathogenesis of COPD by Integrative Bioinformatics Analysis. Int. J. COPD 2019, 14, 2145–2162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Branchfield, K.; Nantie, L.; Verheyden, J.M.; Sui, P.; Wienhold, M.D.; Sun, X. Pulmonary Neuroendocrine Cells Function as Airway Sensors to Control Lung Immune Response. Science 2016, 351, 707–710. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tseng, R.C.; Lee, S.H.; Hsu, H.S.; Chen, B.H.; Tsai, W.C.; Tzao, C.; Wang, Y.C. SLIT2 Attenuation during Lung Cancer Progression Deregulates Beta-Catenin and E-Cadherin and Associates with Poor Prognosis. Cancer Res. 2010, 70, 543–551. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.Y.; Feng, L.; Park, H.T.; Havlioglu, N.; Wen, L.; Tang, H.; Bacon, K.B.; Jiang, Z.; Zhang, X.; Rao, Y. The Neuronal Repellent Slit Inhibits Leukocyte Chemotaxis Induced by Chemotactic Factors. Nature 2001, 410, 948–952. [Google Scholar] [CrossRef]
- Pilling, D.; Chinea, L.E.; Consalvo, K.M.; Gomer, R.H. Different Isoforms of the Neuronal Guidance Molecule Slit2 Directly Cause Chemoattraction or Chemorepulsion of Human Neutrophils. J. Immunol. 2019, 202, 239–248. [Google Scholar] [CrossRef] [Green Version]
- Ye, B.Q.; Geng, Z.H.; Ma, L.; Geng, J.G. Slit2 Regulates Attractive Eosinophil and Repulsive Neutrophil Chemotaxis through Differential SrGAP1 Expression during Lung Inflammation. J. Immunol. 2010, 185, 6294–6305. [Google Scholar] [CrossRef] [Green Version]
- Pilling, D.; Zheng, Z.; Vakil, V.; Gomer, R.H. Fibroblasts Secrete Slit2 to Inhibit Fibrocyte Differentiation and Fibrosis. Proc. Natl. Acad. Sci. USA 2014, 111, 18291–18296. [Google Scholar] [CrossRef] [Green Version]
- Gaspar Marques, J.; Lobato, M.; Leiria Pinto, P.; Neuparth, N.; Carreiro Martins, P. Asthma and COPD “Overlap”: A Treatable Trait or Common Several Treatable-Traits? Eur. Ann. Allergy Clin. Immunol. 2020, 52, 148–159. [Google Scholar] [CrossRef] [Green Version]
- Cukic, V.; Lovre, V.; Dragisic, D.; Ustamujic, A. Asthma and Chronic Obstructive Pulmonary Disease (COPD)—Differences and Similarities. Mater. Sociomed. 2012, 24, 100–105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ning, Y.; Sun, Q.; Dong, Y.; Xu, W.; Zhang, W.; Huang, H.; Li, Q. Slit2-N Inhibits PDGF-Induced Migration in Rat Airway Smooth Muscle Cells: WASP and Arp2/3 Involved. Toxicology 2011, 283, 32–40. [Google Scholar] [CrossRef]
- Zuyderduyn, S.; Sukkar, M.B.; Fust, A.; Dhaliwal, S.; Burgess, J.K. Treating Asthma Means Treating Airway Smooth Muscle Cells. Eur. Respir. J. 2008, 32, 265–274. [Google Scholar] [CrossRef] [PubMed]
- Parameswaran, K.; Radford, K.; Fanat, A.; Stephen, J.; Bonnans, C.; Levy, B.D.; Janssen, L.J.; Cox, P.G. Modulation of Human Airway Smooth Muscle Migration by Lipid Mediators and Th-2 Cytokines. Am. J. Respir. Cell Mol. Biol. 2007, 37, 240–247. [Google Scholar] [CrossRef] [PubMed]
- Lutz, S.M.; Cho, M.H.; Young, K.; Hersh, C.P.; Castaldi, P.J.; McDonald, M.L.; Regan, E.; Mattheisen, M.; DeMeo, D.L.; Parker, M.; et al. A Genome-Wide Association Study Identifies Risk Loci for Spirometric Measures among Smokers of European and African Ancestry. BMC Genet. 2015, 16, 138. [Google Scholar] [CrossRef] [Green Version]
- Ding, L.; Abebe, T.; Beyene, J.; Wilke, R.A.; Goldberg, A.; Woo, J.G.; Martin, L.J.; Rothenberg, M.E.; Rao, M.; Hershey, G.K.; et al. Rank-Based Genome-Wide Analysis Reveals the Association of Ryanodine Receptor-2 Gene Variants with Childhood Asthma among Human Populations. Hum. Genom. 2013, 7, 16. [Google Scholar] [CrossRef] [Green Version]
- Sirugo, G.; Williams, S.M.; Tishkoff, S.A. The Missing Diversity in Human Genetic Studies. Cell 2019, 177, 26–31. [Google Scholar] [CrossRef] [Green Version]
- Potaczek, D.P.; Harb, H.; Michel, S.; Alhamwe, B.A.; Renz, H.; Tost, J. Epigenetics and Allergy: From Basic Mechanisms to Clinical Applications. Epigenomics 2017, 9, 539–571. [Google Scholar] [CrossRef]
- Alashkar Alhamwe, B.; Miethe, S.; Pogge von Strandmann, E.; Potaczek, D.P.; Garn, H. Epigenetic Regulation of Airway Epithelium Immune Functions in Asthma. Front. Immunol. 2020, 11, 1747. [Google Scholar] [CrossRef] [PubMed]
- Reddel, H.K.; Taylor, D.R.; Bateman, E.D.; Boulet, L.P.; Boushey, H.A.; Busse, W.W.; Casale, T.B.; Chanez, P.; Enright, P.L.; Gibson, P.G.; et al. An Official American Thoracic Society/European Respiratory Society Statement: Asthma Control and Exacerbations: Standardizing Endpoints for Clinical Asthma Trials and Clinical Practice. Am. J. Respir. Crit. Care Med. 2009, 180, 59–99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tse, S.M.; Gold, D.R.; Sordillo, J.E.; Hoffman, E.B.; Gillman, M.W.; Rifas-Shiman, S.L.; Fuhlbrigge, A.L.; Tantisira, K.G.; Weiss, S.T.; Litonjua, A.A. Diagnostic Accuracy of the Bronchodilator Response in Children. J. Allergy Clin. Immunol. 2013, 132, 554–559.e5. [Google Scholar] [CrossRef] [Green Version]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J.; et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [Green Version]
- Chang, C.C.; Chow, C.C.; Tellier, L.C.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-Generation PLINK: Rising to the Challenge of Larger and Richer Datasets. Gigascience 2015, 4, 7. [Google Scholar] [CrossRef]
- Kang, H.M. EPACTS (Efficient and Parallelizable Association Container Toolbox) 2016. Available online: http://genome.sph.umich.edu/wiki/EPACTS (accessed on 11 April 2019).
- Price, A.L.; Patterson, N.J.; Plenge, R.M.; Weinblatt, M.E.; Shadick, N.A.; Reich, D. Principal Components Analysis Corrects for Stratification in Genome-Wide Association Studies. Nat. Genet. 2006, 38, 904–909. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.H. Gap: Genetic Analysis Package. R Package Version 1.2.2. 2020. Available online: https://CRAN.R-project.org/package=gap (accessed on 24 July 2019).
- Abecasis, G.R.; Auton, A.; Brooks, L.D.; DePristo, M.A.; Durbin, R.M.; Handsaker, R.E.; Kang, H.M.; Marth, G.T.; McVean, G.A. An Integrated Map of Genetic Variation from 1,092 Human Genomes. Nature 2012, 491, 56–65. [Google Scholar]
- Das, S.; Forer, L.; Schonherr, S.; Sidore, C.; Locke, A.E.; Kwong, A.; Vrieze, S.I.; Chew, E.Y.; Levy, S.; McGue, M.; et al. Next-Generation Genotype Imputation Service and Methods. Nat. Genet. 2016, 48, 1284–1287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuo, K.H.M. Multiple Testing in the Context of Gene Discovery in Sickle Cell Disease Using Genome-Wide Association Studies. Genom. Insights 2017, 10, 1178. [Google Scholar] [CrossRef] [Green Version]
- Plummer, M.; Best, N.; Cowles, K.; Vines, K. CODA: Convergence Diagnosis and Output Analysis for MCMC. R News 2006, 6, 7–11. [Google Scholar]
- Ward, L.D.; Kellis, M. HaploReg v4: Systematic Mining of Putative Causal Variants, Cell Types, Regulators and Target Genes for Human Complex Traits and Disease. Nucleic Acids Res. 2016, 44, D877–D881. [Google Scholar] [CrossRef] [PubMed]


| Total | ICS Non- Responders a | ICS Responders b | p-Value | |
|---|---|---|---|---|
| Sample size | 166 | 94 | 72 | - |
| Gender, n (% male) | 98 (59.0) | 59 (62.8) | 39 (54.2) | 0.264 e |
| Mean age ± SD (years) | 10.9 ± 3.4 | 10.7 ± 3.2 | 11.2 ± 3.5 | 0.461 f |
| Lung function | ||||
| Mean basal FEV1 ± SD (%) c | 87.1 ± 14.8 | 91.3 ± 12.7 | 81.6 ± 15.5 | <0.001 f |
| Mean post-treatment FEV1 ± SD (%) d | 93.7 ± 14.4 | 90.1 ± 13.6 | 98.5 ± 14.2 | <0.001 f |
| Mean ΔFEV1 ± SD (%) | 6.7 ± 12.1 | −1.2 ± 7.8 | 16.9 ± 8.7 | <0.001 f |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hernandez-Pacheco, N.; Gorenjak, M.; Li, J.; Repnik, K.; Vijverberg, S.J.; Berce, V.; Jorgensen, A.; Karimi, L.; Schieck, M.; Samedy-Bates, L.-A.; et al. Identification of ROBO2 as a Potential Locus Associated with Inhaled Corticosteroid Response in Childhood Asthma. J. Pers. Med. 2021, 11, 733. https://doi.org/10.3390/jpm11080733
Hernandez-Pacheco N, Gorenjak M, Li J, Repnik K, Vijverberg SJ, Berce V, Jorgensen A, Karimi L, Schieck M, Samedy-Bates L-A, et al. Identification of ROBO2 as a Potential Locus Associated with Inhaled Corticosteroid Response in Childhood Asthma. Journal of Personalized Medicine. 2021; 11(8):733. https://doi.org/10.3390/jpm11080733
Chicago/Turabian StyleHernandez-Pacheco, Natalia, Mario Gorenjak, Jiang Li, Katja Repnik, Susanne J. Vijverberg, Vojko Berce, Andrea Jorgensen, Leila Karimi, Maximilian Schieck, Lesly-Anne Samedy-Bates, and et al. 2021. "Identification of ROBO2 as a Potential Locus Associated with Inhaled Corticosteroid Response in Childhood Asthma" Journal of Personalized Medicine 11, no. 8: 733. https://doi.org/10.3390/jpm11080733