Whole Exome Sequencing in Healthy Individuals of Extreme Constitution Types Reveals Differential Disease Risk: A Novel Approach towards Predictive Medicine
Abstract
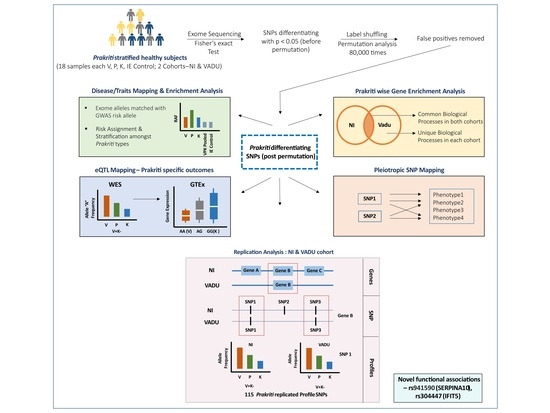
:1. Introduction
2. Materials and Methods
2.1. Sample Description
2.2. Whole Exome Sequencing (WES) and Variant Calling
2.3. Mapping and Enrichment Analysis for Disease/Traits Associations
2.4. Multi-System Phenotype Association Analysis Using GeneAtlas
2.5. Replication Analysis of Prakriti Differentiating SNPs
2.6. Power Analysis
3. Results
3.1. Genetic Differences amongst Healthy Prakriti Types Remain Significant after Permutation Analysis
3.2. Distinct Enrichment of Biological Processes in Prakriti Groups: Similar Patterns across Both Cohorts
3.3. Significant Enrichment of Prakriti Differentiating SNPs for Variants with Common and Complex Diseases
3.4. Enriched Disease/Traits Associated with Prakriti Differentiating Genetic Variations Enable Risk Stratification
3.5. Risk Stratification amongst Healthy Individuals: Potential for Early Identification
3.6. Multi-System Phenotypic Associations of Prakriti Differentiating Variants in GeneATLAS
3.7. Similar Patterns of Exonic Differences: Identification of Prakriti Replicated Profile SNPs across Both Cohorts
3.8. Prakriti Replicated Profile SNPs Significantly Differ from Background Population
3.9. Novel Leads from Prakriti Replicated Profile SNPs Confer Differential Disease Trajectories
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Topol, E.J. Individualized Medicine from Prewomb to Tomb. Cell 2014, 157, 241–253. [Google Scholar] [CrossRef] [Green Version]
- Snyder, M.; Weissman, S.; Gerstein, M. Personal Phenotypes to Go with Personal Genomes. Mol. Syst. Biol. 2009, 5, 273. [Google Scholar] [CrossRef] [PubMed]
- Loscalzo, J.; Barabasi, A.-L. Systems Biology and the Future of Medicine. Wiley Interdiscip. Rev. Syst. Biol. Med. 2011, 3, 619–627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, R.; Snyder, M. Promise of Personalized Omics to Precision Medicine. Wiley Interdiscip. Rev. Syst. Biol. Med. 2013, 5, 73–82. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Price, N.D.; Magis, A.T.; Earls, J.C.; Glusman, G.; Levy, R.; Lausted, C.; McDonald, D.T.; Kusebauch, U.; Moss, C.L.; Zhou, Y.; et al. A Wellness Study of 108 Individuals Using Personal, Dense, Dynamic Data Clouds. Nat. Biotechnol. 2017, 35, 747–756. [Google Scholar] [CrossRef]
- Hou, Y.-C.C.; Yu, H.-C.; Martin, R.; Cirulli, E.T.; Schenker-Ahmed, N.M.; Hicks, M.; Cohen, I.V.; Jönsson, T.J.; Heister, R.; Napier, L.; et al. Precision Medicine Integrating Whole-Genome Sequencing, Comprehensive Metabolomics, and Advanced Imaging. Proc. Natl. Acad. Sci. USA 2020, 117, 3053–3062. [Google Scholar] [CrossRef] [Green Version]
- Lemonnier, N.; Zhou, G.-B.; Prasher, B.; Mukerji, M.; Chen, Z.; Brahmachari, S.K.; Noble, D.; Auffray, C.; Sagner, M. Traditional Knowledge-Based Medicine: A Review of History, Principles, and Relevance in the Present Context of P4 Systems Medicine. Prog. Prev. Med. 2017, 2, e0011. [Google Scholar] [CrossRef]
- Tam, V.; Patel, N.; Turcotte, M.; Bossé, Y.; Paré, G.; Meyre, D. Benefits and Limitations of Genome-Wide Association Studies. Nat. Rev. Genet. 2019, 20, 467–484. [Google Scholar] [CrossRef]
- Goh, K.-I.; Cusick, M.E.; Valle, D.; Childs, B.; Vidal, M.; Barabási, A.-L. The Human Disease Network. Proc. Natl. Acad. Sci. USA 2007, 104, 8685–8690. [Google Scholar] [CrossRef] [Green Version]
- Welter, D.; MacArthur, J.; Morales, J.; Burdett, T.; Hall, P.; Junkins, H.; Klemm, A.; Flicek, P.; Manolio, T.; Hindorff, L.; et al. The NHGRI GWAS Catalog, a Curated Resource of SNP-Trait Associations. Nucleic Acids Res. 2014, 42, D1001–D1006. [Google Scholar] [CrossRef]
- Bush, W.S.; Oetjens, M.T.; Crawford, D.C. Unravelling the Human Genome-Phenome Relationship Using Phenome-Wide Association Studies. Nat. Rev. Genet. 2016, 17, 129–145. [Google Scholar] [CrossRef] [PubMed]
- Denny, J.C.; Bastarache, L.; Ritchie, M.D.; Carroll, R.J.; Zink, R.; Mosley, J.D.; Field, J.R.; Pulley, J.M.; Ramirez, A.H.; Bowton, E.; et al. Systematic Comparison of Phenome-Wide Association Study of Electronic Medical Record Data and Genome-Wide Association Study Data. Nat. Biotechnol. 2013, 31, 1102–1110. [Google Scholar] [CrossRef] [Green Version]
- Millard, L.A.C.; Davies, N.M.; Tilling, K.; Gaunt, T.R.; Davey Smith, G. Searching for the Causal Effects of Body Mass Index in over 300 000 Participants in UK Biobank, Using Mendelian Randomization. PLoS Genet. 2019, 15, e1007951. [Google Scholar] [CrossRef] [Green Version]
- Verma, A.; Lucas, A.; Verma, S.S.; Zhang, Y.; Josyula, N.; Khan, A.; Hartzel, D.N.; Lavage, D.R.; Leader, J.; Ritchie, M.D.; et al. PheWAS and Beyond: The Landscape of Associations with Medical Diagnoses and Clinical Measures across 38,662 Individuals from Geisinger. Am. J. Hum. Genet. 2018, 102, 592–608. [Google Scholar] [CrossRef] [Green Version]
- Phenome-Wide Association Study (PheWAS) for Detection of Pleiotropy within the Population Architecture Using Genomics and Epidemiology (PAGE) Network—PubMed. Available online: https://pubmed.ncbi.nlm.nih.gov/23382687/ (accessed on 31 October 2021).
- Cai, T.; Zhang, Y.; Ho, Y.-L.; Link, N.; Sun, J.; Huang, J.; Cai, T.A.; Damrauer, S.; Ahuja, Y.; Honerlaw, J.; et al. Association of Interleukin 6 Receptor Variant With Cardiovascular Disease Effects of Interleukin 6 Receptor Blocking Therapy: A Phenome-Wide Association Study. JAMA Cardiol. 2018, 3, 849–857. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rastegar-Mojarad, M.; Ye, Z.; Kolesar, J.M.; Hebbring, S.J.; Lin, S.M. Opportunities for Drug Repositioning from Phenome-Wide Association Studies. Nat. Biotechnol. 2015, 33, 342–345. [Google Scholar] [CrossRef]
- Diogo, D.; Tian, C.; Franklin, C.S.; Alanne-Kinnunen, M.; March, M.; Spencer, C.C.A.; Vangjeli, C.; Weale, M.E.; Mattsson, H.; Kilpeläinen, E.; et al. Phenome-Wide Association Studies across Large Population Cohorts Support Drug Target Validation. Nat. Commun. 2018, 9, 4285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, W.; Gao, C.; Xu, Y.; Li, B.; Ruderfer, D.M.; Chen, Y. Learning Opportunities for Drug Repositioning via GWAS and PheWAS Findings. AMIA Jt. Summits Transl. Sci. Proc. 2018, 2017, 237–246. [Google Scholar] [PubMed]
- Hebbring, S.J. The Challenges, Advantages and Future of Phenome-Wide Association Studies. Immunology 2014, 141, 157–165. [Google Scholar] [CrossRef]
- Ried, J.S.; Jeff, M.J.; Chu, A.Y.; Bragg-Gresham, J.L.; van Dongen, J.; Huffman, J.E.; Ahluwalia, T.S.; Cadby, G.; Eklund, N.; Eriksson, J.; et al. A Principal Component Meta-Analysis on Multiple Anthropometric Traits Identifies Novel Loci for Body Shape. Nat. Commun. 2016, 7, 13357. [Google Scholar] [CrossRef]
- Emond, M.J.; Louie, T.; Emerson, J.; Zhao, W.; Mathias, R.A.; Knowles, M.R.; Wright, F.A.; Rieder, M.J.; Tabor, H.K.; Nickerson, D.A.; et al. Exome Sequencing of Extreme Phenotypes Identifies DCTN4 as a Modifier of Chronic Pseudomonas aeruginosa Infection in Cystic Fibrosis. Nat. Genet. 2012, 44, 886–889. [Google Scholar] [CrossRef] [PubMed]
- Emond, M.J.; Louie, T.; Emerson, J.; Chong, J.X.; Mathias, R.A.; Knowles, M.R.; Rieder, M.J.; Tabor, H.K.; Nickerson, D.A.; Barnes, K.C.; et al. Exome Sequencing of Phenotypic Extremes Identifies CAV2 and TMC6 as Interacting Modifiers of Chronic Pseudomonas aeruginosa Infection in Cystic Fibrosis. PLoS Genet. 2015, 11, e1005273. [Google Scholar] [CrossRef] [Green Version]
- Aubart, M.; Gazal, S.; Arnaud, P.; Benarroch, L.; Gross, M.-S.; Buratti, J.; Boland, A.; Meyer, V.; Zouali, H.; Hanna, N.; et al. Association of Modifiers and Other Genetic Factors Explain Marfan Syndrome Clinical Variability. Eur. J. Hum. Genet. 2018, 26, 1759–1772. [Google Scholar] [CrossRef] [Green Version]
- Fay, A.P.; de Velasco, G.; Ho, T.H.; Van Allen, E.M.; Murray, B.; Albiges, L.; Signoretti, S.; Hakimi, A.A.; Stanton, M.L.; Bellmunt, J.; et al. Whole-Exome Sequencing in Two Extreme Phenotypes of Response to VEGF-Targeted Therapies in Patients With Metastatic Clear Cell Renal Cell Carcinoma. J. Natl. Compr. Canc. Netw. 2016, 14, 820–824. [Google Scholar] [CrossRef] [Green Version]
- Chan, Y.; Holmen, O.L.; Dauber, A.; Vatten, L.; Havulinna, A.S.; Skorpen, F.; Kvaløy, K.; Silander, K.; Nguyen, T.T.; Willer, C.; et al. Common Variants Show Predicted Polygenic Effects on Height in the Tails of the Distribution, except in Extremely Short Individuals. PLoS Genet. 2011, 7, e1002439. [Google Scholar] [CrossRef] [Green Version]
- Naslavsky, M.S.; Vidigal, M.; Matos, L.d.R.B.; Cória, V.R.; Batista Junior, P.B.; Razuk, Á.; Saldiva, P.H.N.; Dolhnikoff, M.; Schidlowski, L.; Prando, C.; et al. Extreme Phenotypes Approach to Investigate Host Genetics and COVID-19 Outcomes. Genet. Mol. Biol. 2021, 44, e20200302. [Google Scholar] [CrossRef] [PubMed]
- Dueker, N.D.; Doliner, B.; Gardener, H.; Dong, C.; Beecham, A.; Della-Morte, D.; Sacco, R.L.; Blanton, S.H.; Wang, L.; Rundek, T. Extreme Phenotype Approach Suggests Taste Transduction Pathway for Carotid Plaque in a Multi-Ethnic Cohort. Stroke 2020, 51, 2761–2769. [Google Scholar] [CrossRef] [PubMed]
- Sethi, T.P.; Prasher, B.; Mukerji, M. Ayurgenomics: A New Way of Threading Molecular Variability for Stratified Medicine. ACS Chem. Biol. 2011, 6, 875–880. [Google Scholar] [CrossRef]
- Sagner, M.; McNeil, A.; Puska, P.; Auffray, C.; Price, N.D.; Hood, L.; Lavie, C.J.; Han, Z.-G.; Chen, Z.; Brahmachari, S.K.; et al. The P4 Health Spectrum—A Predictive, Preventive, Personalized and Participatory Continuum for Promoting Healthspan. Prog. Cardiovasc. Dis. 2017, 59, 506–521. [Google Scholar] [CrossRef] [Green Version]
- Prasher, B.; Gibson, G.; Mukerji, M. Genomic Insights into Ayurvedic and Western Approaches to Personalized Medicine. J. Genet. 2016, 95, 209–228. [Google Scholar] [CrossRef]
- Tiwari, P.; Kutum, R.; Sethi, T.; Shrivastava, A.; Girase, B.; Aggarwal, S.; Patil, R.; Agarwal, D.; Gautam, P.; Agrawal, A.; et al. Recapitulation of Ayurveda Constitution Types by Machine Learning of Phenotypic Traits. PLoS ONE 2017, 12, e0185380. [Google Scholar] [CrossRef] [PubMed]
- Prasher, B.; Negi, S.; Aggarwal, S.; Mandal, A.K.; Sethi, T.P.; Deshmukh, S.R.; Purohit, S.G.; Sengupta, S.; Khanna, S.; Mohammad, F.; et al. Whole Genome Expression and Biochemical Correlates of Extreme Constitutional Types Defined in Ayurveda. J. Transl. Med. 2008, 6, 48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Govindaraj, P.; Nizamuddin, S.; Sharath, A.; Jyothi, V.; Rotti, H.; Raval, R.; Nayak, J.; Bhat, B.K.; Prasanna, B.V.; Shintre, P.; et al. Genome-Wide Analysis Correlates Ayurveda Prakriti. Sci. Rep. 2015, 5, 15786. Available online: https://www.nature.com/articles/srep15786 (accessed on 17 November 2021). [CrossRef] [PubMed] [Green Version]
- Rotti, H.; Mallya, S.; Kabekkodu, S.P.; Chakrabarty, S.; Bhale, S.; Bharadwaj, R.; Bhat, B.K.; Dedge, A.P.; Dhumal, V.R.; Gangadharan, G.G.; et al. DNA Methylation Analysis of Phenotype Specific Stratified Indian Population. J. Transl. Med. 2015, 13, 151. Available online: https://translational-medicine.biomedcentral.com/articles/10.1186/s12967-015-0506-0 (accessed on 17 November 2021). [CrossRef] [Green Version]
- Aggarwal, S.; Negi, S.; Jha, P.; Singh, P.K.; Stobdan, T.; Pasha, M.A.Q.; Ghosh, S.; Agrawal, A.; Prasher, B.; Indian Genome Variation Consortium; et al. EGLN1 Involvement in High-Altitude Adaptation Revealed through Genetic Analysis of Extreme Constitution Types Defined in Ayurveda. Proc. Natl. Acad. Sci. USA 2010, 107, 18961–18966. [Google Scholar] [CrossRef] [Green Version]
- Chakraborty, S.; Singhmar, S.; Singh, D.; Maulik, M.; Patil, R.; Agrawal, S.K.; Mishra, A.; Ghazi, M.; Vats, A.; Natarajan, V.T.; et al. Baseline Cell Proliferation Rates and Response to UV Differ in Lymphoblastoid Cell Lines Derived from Healthy Individuals of Extreme Constitution Types. Cell Cycle 2021, 20, 903–913. [Google Scholar] [CrossRef]
- Aggarwal, S.; Gheware, A.; Agrawal, A.; Ghosh, S.; Prasher, B.; Mukerji, M. Indian Genome Variation Consortium Combined Genetic Effects of EGLN1 and VWF Modulate Thrombotic Outcome in Hypoxia Revealed by Ayurgenomics Approach. J. Transl. Med. 2015, 13, 184. [Google Scholar] [CrossRef] [Green Version]
- Prasher, B.; Varma, B.; Kumar, A.; Khuntia, B.K.; Pandey, R.; Narang, A.; Tiwari, P.; Kutum, R.; Guin, D.; Kukreti, R.; et al. Ayurgenomics for Stratified Medicine: TRISUTRA Consortium Initiative across Ethnically and Geographically Diverse Indian Populations. J. Ethnopharmacol. 2017, 197, 274–293. [Google Scholar] [CrossRef]
- Indian Genome Variation Consortium The Indian Genome Variation Database (IGVdb): A Project Overview. Hum. Genet. 2005, 118, 1–11. [CrossRef]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional Annotation of Genetic Variants from High-Throughput Sequencing Data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef]
- Zhao, H.; Sun, Z.; Wang, J.; Huang, H.; Kocher, J.-P.; Wang, L. CrossMap: A Versatile Tool for Coordinate Conversion between Genome Assemblies. Bioinformatics 2014, 30, 1006–1007. [Google Scholar] [CrossRef]
- Canela-Xandri, O.; Rawlik, K.; Tenesa, A. An Atlas of Genetic Associations in UK Biobank. Nat. Genet. 2018, 50, 1593–1599. [Google Scholar] [CrossRef] [PubMed]
- Giner, G.; Smyth, G.K. Statmod: Probability Calculations for the Inverse Gaussian Distribution. R J. 2016, 8, 339. [Google Scholar] [CrossRef]
- Zhang, B.; Liu, X.; Chen, W.; Chen, L. IFIT5 Potentiates Anti-Viral Response through Enhancing Innate Immune Signaling Pathways. Acta Biochim. Biophys. Sin. (Shanghai) 2013, 45, 867–874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rau, J.C.; Beaulieu, L.M.; Huntington, J.A.; Church, F.C. Serpins in Thrombosis, Hemostasis and Fibrinolysis. J. Thromb. Haemost. 2007, 5, 102–115. [Google Scholar] [CrossRef] [PubMed]
- Yao, C.; Chen, G.; Song, C.; Keefe, J.; Mendelson, M.; Huan, T.; Sun, B.B.; Laser, A.; Maranville, J.C.; Wu, H.; et al. Genome-wide Mapping of Plasma Protein QTLs Identifies Putatively Causal Genes and Pathways for Cardiovascular Disease. Nat. Commun. 2018, 9, 3268. Available online: https://www.nature.com/articles/s41467-018-05512-x (accessed on 18 November 2021). [CrossRef]
- BasuRay, S.; Wang, Y.; Smagris, E.; Cohen, J.C.; Hobbs, H.H. Accumulation of PNPLA3 on Lipid Droplets Is the Basis of Associated Hepatic Steatosis. Proc. Natl. Acad. Sci. USA 2019, 116, 9521–9526. [Google Scholar] [CrossRef] [Green Version]
- Kipper, S.; Hamad, S.; Caly, L.; Avrahami, D.; Bacharach, E.; Jans, D.A.; Gerber, D.; Bajorek, M. New Host Factors Important for Respiratory Syncytial Virus (RSV) Replication Revealed by a Novel Microfluidics Screen for Interactors of Matrix (M) Protein. Mol. Cell. Proteom. 2015, 14, 532–543. [Google Scholar] [CrossRef] [Green Version]
- Corral, J.; González-Conejero, R.; Soria, J.M.; González-Porras, J.R.; Pérez-Ceballos, E.; Lecumberri, R.; Roldán, V.; Souto, J.C.; Miñano, A.; Hernández-Espinosa, D.; et al. A Nonsense Polymorphism in the Protein Z-Dependent Protease Inhibitor Increases the Risk for Venous Thrombosis. Blood 2006, 108, 177–183. [Google Scholar] [CrossRef] [Green Version]
- Juyal, R.C.; Negi, S.; Wakhode, P.; Bhat, S.; Bhat, B.; Thelma, B.K. Potential of Ayurgenomics Approach in Complex Trait Research: Leads from a Pilot Study on Rheumatoid Arthritis. PLoS ONE 2012, 7, e45752. [Google Scholar] [CrossRef] [Green Version]





| Cohort | SNP | Gene | GWAS Disease/Trait | Risk Allele | Risk Allele Frequency (RAF) | Differentiating Prakriti Groups | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| V | P | K | VPK Pooled | IE Control | GWAS | ||||||
| Vadu | rs11603334 (5′UTR variation) | ARAP1 | Fasting Blood Proinsulin levels | A | 0.08 * | 0.25 | 0.33 ## | 0.22 | 0.12 | 0.25 | V vs. C, V vs. K |
| rs1552224 | ARAP1 | Acute insulin response | A | 0.92 ## | 0.72 | 0.67 * | 0.77 | 0.88 | NR | V vs. K, K vs. C | |
| rs3014246 (missense) | CCDC17 | Apolipoprotein A1 levels | C | 0.5 ## | 0.39 | 0.19 * | 0.36 | 0.50 | 0.29 | V vs. K, K vs. C | |
| rs682331 (3′UTR variation) | NIBAN1 | Obesity related traits | G | 0.69 ## | 0.27 | 0.2 * | 0.39 | 0.41 | 0.44 | V vs. C, V vs. P, V vs. K | |
| rs3811445 (synonymous) | TRIM58 | Immature fraction of reticulocytes | G | 0.58 | 0.79 ## | 0.39 * | 0.58 | 0.68 | 0.58 | P vs. K, K vs. C | |
| rs10922162 | ASPM | End-stage coagulation | C | 0.72 | 0.56 * | 0.81 ## | 0.7 | 0.85 | 0.83 | P vs. K, P vs. C | |
| rs1801222 | CUBN | Homocysteine levels | A | 0.31 ## | 0.03 * | 0.24 ## | 0.19 | 0.09 | 0.34 | P vs. K, V vs. P, V vs. C | |
| rs257377 | PRKAR2B | LDL cholesterol | G | 0.75 * | 0.83 | 0.97 ## | 0.85 | 0.71 | 0.79 | V vs. K, K vs. C | |
| rs738409 (missense) | PNPLA3 | Cirrhosis | G | 0.28 | 0.08 * | 0.36 ## | 0.24 | 0.09 | 0.27 | K vs. C, P vs. K | |
| Hb conc | 0.21 | ||||||||||
| Hb conc | 0.26 | ||||||||||
| Liver enzymes level | 0.23 | ||||||||||
| Liver fibrosis | 0.21 | ||||||||||
| Red cell distribution width | 0.21 | ||||||||||
| Total triglyceride levels | 0.36 | ||||||||||
| T2D | 0.22 | ||||||||||
| NI | rs699 (nonsynonymous) | AGT | Mean Arterial Pressure | A | 0.36 ## | 0.25 | 0.11 * | 0.24 | 0.38 | 0.48 | V vs. K, K vs. C |
| rs2792751 (nonsynonymous) | GPAM | HDL Cholesterol levels, Apolipoprotein A1 levels | T | 0.16 | 0.37 ## | 0.04 * | 0.19 | 0.11 | 0.27 | P vs. K, P vs. C | |
| rs3764002 (nonsynonymous) | WSCD2 | T2D, Waist-to-hip ratio | C | 0.83 ## | 0.64 | 0.56 * | 0.68 | 0.58 | 0.72,0.73 | V vs. K, V vs. C | |
| rs3764002 (nonsynonymous) | WSCD2 | Risk taking tendency, Predicted visceral adipose tissue | T | 0.17 * | 0.36 | 0.44 ## | 0.32 | 0.41 | 0.26 | V vs. K,V vs. C | |
| rs10793625 (5′UTR variant) | WASH2C | Mean corpuscular Hb levels | C | 0.67 * | 0.81 | 0.94 ## | 0.81 | 0.61 | 0.79 | V vs. K, K vs. C | |
| rs675531 (nonsynonymous) | THEMIS | Recalcitrant atopic dermatitis | C | 0.43 | 0.66 ## | 0.33 * | 0.47 | 0.30 | 0.11 | P vs. K, P vs. C | |
| rs8073060 (missense) | SLFN14 | Platelet count | A | 0.15 * | 0.44 ## | 0.35 | 0.31 | 0.44 | 0.29 | V vs. P, V vs. C | |
| rs2073498 (missense) | RASSF1 | Feeling worry | A | 0.14 | 0.25 ## | 0.06 * | 0.15 | 0.05 | 0.11 | P vs. K, P vs. C | |
| rs41269255 (nonsynonymous) | POM121L2 | Depressive symptoms | T | 0 * | 0.08 | 0.21 ## | 0.1 | 0.02 | 0.11 | V vs. K, K vs. C | |
| rs17412833 (nonsynonymous) | HLA-DQB1 | Lactate dehydrogenase levels | T | 0.2 * | 0.53 ## | 0.38 | 0.37 | 0.52 | 0.13 | V vs. P, V vs. C | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abbas, T.; Chaturvedi, G.; Prakrithi, P.; Pathak, A.K.; Kutum, R.; Dakle, P.; Narang, A.; Manchanda, V.; Patil, R.; Aggarwal, D.; et al. Whole Exome Sequencing in Healthy Individuals of Extreme Constitution Types Reveals Differential Disease Risk: A Novel Approach towards Predictive Medicine. J. Pers. Med. 2022, 12, 489. https://doi.org/10.3390/jpm12030489
Abbas T, Chaturvedi G, Prakrithi P, Pathak AK, Kutum R, Dakle P, Narang A, Manchanda V, Patil R, Aggarwal D, et al. Whole Exome Sequencing in Healthy Individuals of Extreme Constitution Types Reveals Differential Disease Risk: A Novel Approach towards Predictive Medicine. Journal of Personalized Medicine. 2022; 12(3):489. https://doi.org/10.3390/jpm12030489
Chicago/Turabian StyleAbbas, Tahseen, Gaura Chaturvedi, P. Prakrithi, Ankit Kumar Pathak, Rintu Kutum, Pushkar Dakle, Ankita Narang, Vijeta Manchanda, Rutuja Patil, Dhiraj Aggarwal, and et al. 2022. "Whole Exome Sequencing in Healthy Individuals of Extreme Constitution Types Reveals Differential Disease Risk: A Novel Approach towards Predictive Medicine" Journal of Personalized Medicine 12, no. 3: 489. https://doi.org/10.3390/jpm12030489