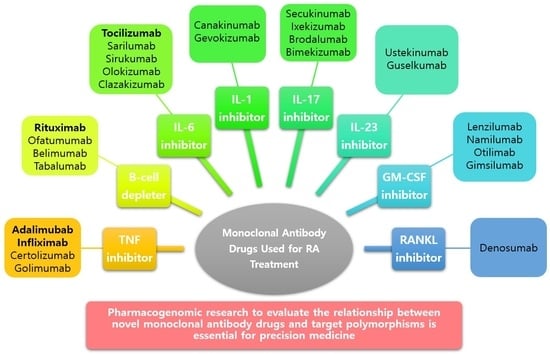
Pharmacogenomics of Monoclonal Antibodies for the Treatment of Rheumatoid Arthritis
Abstract
:1. Introduction
2. Adalimumab
2.1. FCGR2A
2.2. DHX32
2.3. RGS12
2.4. IL-6
2.5. PTPN22
2.6. TNF
2.7. TNFR2
3. Infliximab
3.1. FCGR2A and FCGR3A
3.2. RGS12
3.3. PTPN22
3.4. MHC
3.5. TNF
3.6. TNFR1 and TNFR2
4. Rituximab
4.1. FCGR2A and FCGR3
4.2. BAFF
4.3. IL-6
4.4. TGFβ1
5. Tocilizumab
5.1. FCGR2A and FCGR3A
5.2. HLA-DRB1
5.3. IL-6R
6. Newer Monoclonal Antibody Drugs and Genetic Polymorphisms of Their Targets
7. Conclusions and Future Challenges
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jameson, J.L.; Longo, D.L. Precision Medicine—Personalized, Problematic, and Promising. N. Engl. J. Med. 2015, 372, 2229–2234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- König, I.R.; Fuchs, O.; Hansen, G.; von Mutius, E.; Kopp, M.V. What Is Precision Medicine? Eur. Respir. J. 2017, 50, 1700391. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kohler, S. Precision Medicine—Moving Away from One-Size-Fits-All. Quest 2018, 14, 12–15. [Google Scholar] [CrossRef]
- McDonald, E.S.; Clark, A.S.; Tchou, J.; Zhang, P.; Freedman, G.M. Clinical Diagnosis and Management of Breast Cancer. J. Nucl. Med. 2016, 57, 9S–16S. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Sanzo, M.; Cipolloni, L.; Borro, M.; La Russa, R.; Santurro, A.; Scopetti, M.; Simmaco, M.; Frati, P. Clinical Applications of Personalized Medicine: A New Paradigm and Challenge. Curr. Pharm. Biotechnol. 2017, 18, 194–203. [Google Scholar] [CrossRef]
- van der Wouden, C.H.; Böhringer, S.; Cecchin, E.; Cheung, K.-C.; Dávila-Fajardo, C.L.; Deneer, V.H.M.; Dolžan, V.; Ingelman-Sundberg, M.; Jönsson, S.; Karlsson, M.O.; et al. Generating Evidence for Precision Medicine: Considerations Made by the Ubiquitous Pharmacogenomics Consortium When Designing and Operationalizing the PREPARE Study. Pharm. Genom. 2020, 30, 131–144. [Google Scholar] [CrossRef]
- Cecchin, E.; Stocco, G. Pharmacogenomics and Personalized Medicine. Genes 2020, 11, 679. [Google Scholar] [CrossRef] [PubMed]
- Weinshilboum, R.M.; Wang, L. Pharmacogenomics: Precision Medicine and Drug Response. Mayo Clin. Proc. 2017, 92, 1711–1722. [Google Scholar] [CrossRef] [PubMed]
- Chenoweth, M.J.; Giacomini, K.M.; Pirmohamed, M.; Hill, S.L.; van Schaik, R.H.N.; Schwab, M.; Shuldiner, A.R.; Relling, M.V.; Tyndale, R.F. Global Pharmacogenomics Within Precision Medicine: Challenges and Opportunities. Clin. Pharmacol. Ther. 2020, 107, 57–61. [Google Scholar] [CrossRef] [PubMed]
- Kesik-Brodacka, M. Progress in Biopharmaceutical Development. Biotechnol. Appl. Biochem. 2018, 65, 306–322. [Google Scholar] [CrossRef] [Green Version]
- Mitragotri, S.; Burke, P.A.; Langer, R. Overcoming the Challenges in Administering Biopharmaceuticals: Formulation and Delivery Strategies. Nat. Rev. Drug Discov. 2014, 13, 655–672. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Minghetti, P.; Rocco, P.; Cilurzo, F.; Vecchio, L.D.; Locatelli, F. The Regulatory Framework of Biosimilars in the European Union. Drug Discov. Today 2012, 17, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Sivaccumar, J.; Sandomenico, A.; Vitagliano, L.; Ruvo, M. Monoclonal Antibodies: A Prospective and Retrospective View. Curr. Med. Chem. 2021, 28, 435–471. [Google Scholar] [CrossRef]
- Walsh, G. Biopharmaceutical Benchmarks 2018. Nat. Biotechnol. 2018, 36, 1136–1145. [Google Scholar] [CrossRef]
- Smolen, J.S.; Aletaha, D. Rheumatoid Arthritis Therapy Reappraisal: Strategies, Opportunities and Challenges. Nat. Rev. Rheumatol. 2015, 11, 276–289. [Google Scholar] [CrossRef] [PubMed]
- Smolen, J.S.; Aletaha, D.; McInnes, I.B. Rheumatoid Arthritis. Lancet 2016, 388, 2023–2038. [Google Scholar] [CrossRef]
- Kirwan, J.R. The Effect of Glucocorticoids on Joint Destruction in Rheumatoid Arthritis. The Arthritis and Rheumatism Council Low-Dose Glucocorticoid Study Group. N. Engl. J. Med. 1995, 333, 142–146. [Google Scholar] [CrossRef] [PubMed]
- Donahue, K.E.; Gartlehner, G.; Jonas, D.E.; Lux, L.J.; Thieda, P.; Jonas, B.L.; Hansen, R.A.; Morgan, L.C.; Lohr, K.N. Systematic Review: Comparative Effectiveness and Harms of Disease-Modifying Medications for Rheumatoid Arthritis. Ann. Intern. Med. 2008, 148, 124–134. [Google Scholar] [CrossRef]
- Cho, S.-K.; Sung, Y.-K. Treatment strategy for patients with rheumatoid arthritis. J. Korean Med. Assoc. 2020, 63, 422–430. [Google Scholar] [CrossRef]
- Burmester, G.R.; Pope, J.E. Novel Treatment Strategies in Rheumatoid Arthritis. Lancet 2017, 389, 2338–2348. [Google Scholar] [CrossRef]
- Schoels, M.; Aletaha, D.; Smolen, J.S.; Wong, J.B. Comparative Effectiveness and Safety of Biological Treatment Options after Tumour Necrosis Factor α Inhibitor Failure in Rheumatoid Arthritis: Systematic Review and Indirect Pairwise Meta-Analysis. Ann. Rheum. Dis. 2012, 71, 1303–1308. [Google Scholar] [CrossRef] [PubMed]
- Abbasi, M.; Mousavi, M.J.; Jamalzehi, S.; Alimohammadi, R.; Bezvan, M.H.; Mohammadi, H.; Aslani, S. Strategies toward Rheumatoid Arthritis Therapy; the Old and the New. J. Cell. Physiol. 2019, 234, 10018–10031. [Google Scholar] [CrossRef] [PubMed]
- Radner, H.; Aletaha, D. Anti-TNF in Rheumatoid Arthritis: An Overview. Wien. Med. Wochenschr. 2015, 165, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Saxne, T.; Palladino, M.A.; Heinegård, D.; Talal, N.; Wollheim, F.A. Detection of Tumor Necrosis Factor Alpha but Not Tumor Necrosis Factor Beta in Rheumatoid Arthritis Synovial Fluid and Serum. Arthritis Rheum. 1988, 31, 1041–1045. [Google Scholar] [CrossRef] [PubMed]
- Feldmann, M.; Maini, R.N. Lasker Clinical Medical Research Award. TNF Defined as a Therapeutic Target for Rheumatoid Arthritis and Other Autoimmune Diseases. Nat. Med. 2003, 9, 1245–1250. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.-J.; Anzaghe, M.; Schülke, S. Update on the Pathomechanism, Diagnosis, and Treatment Options for Rheumatoid Arthritis. Cells 2020, 9, 880. [Google Scholar] [CrossRef] [Green Version]
- Bertolini, D.R.; Nedwin, G.E.; Bringman, T.S.; Smith, D.D.; Mundy, G.R. Stimulation of Bone Resorption and Inhibition of Bone Formation in Vitro by Human Tumour Necrosis Factors. Nature 1986, 319, 516–518. [Google Scholar] [CrossRef]
- Brennan, F.M.; McInnes, I.B. Evidence That Cytokines Play a Role in Rheumatoid Arthritis. J. Clin. Investig. 2008, 118, 3537–3545. [Google Scholar] [CrossRef] [Green Version]
- Osta, B.; Benedetti, G.; Miossec, P. Classical and Paradoxical Effects of TNF-α on Bone Homeostasis. Front. Immunol. 2014, 5, 48. [Google Scholar] [CrossRef]
- Lipsky, P.E.; van der Heijde, D.M.; St Clair, E.W.; Furst, D.E.; Breedveld, F.C.; Kalden, J.R.; Smolen, J.S.; Weisman, M.; Emery, P.; Feldmann, M.; et al. Infliximab and Methotrexate in the Treatment of Rheumatoid Arthritis. Anti-Tumor Necrosis Factor Trial in Rheumatoid Arthritis with Concomitant Therapy Study Group. N. Engl. J. Med. 2000, 343, 1594–1602. [Google Scholar] [CrossRef] [Green Version]
- Weinblatt, M.E.; Keystone, E.C.; Furst, D.E.; Moreland, L.W.; Weisman, M.H.; Birbara, C.A.; Teoh, L.A.; Fischkoff, S.A.; Chartash, E.K. Adalimumab, a Fully Human Anti-Tumor Necrosis Factor Alpha Monoclonal Antibody, for the Treatment of Rheumatoid Arthritis in Patients Taking Concomitant Methotrexate: The ARMADA Trial. Arthritis Rheum. 2003, 48, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Elliott, M.J.; Maini, R.N.; Feldmann, M.; Kalden, J.R.; Antoni, C.; Smolen, J.S.; Leeb, B.; Breedveld, F.C.; Macfarlane, J.D.; Bijl, H. Randomised Double-Blind Comparison of Chimeric Monoclonal Antibody to Tumour Necrosis Factor Alpha (CA2) versus Placebo in Rheumatoid Arthritis. Lancet 1994, 344, 1105–1110. [Google Scholar] [CrossRef]
- Moreland, L.W.; Baumgartner, S.W.; Schiff, M.H.; Tindall, E.A.; Fleischmann, R.M.; Weaver, A.L.; Ettlinger, R.E.; Cohen, S.; Koopman, W.J.; Mohler, K.; et al. Treatment of Rheumatoid Arthritis with a Recombinant Human Tumor Necrosis Factor Receptor (P75)-Fc Fusion Protein. N. Engl. J. Med. 1997, 337, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Szekanecz, Z.; Besenyei, T.; Paragh, G.; Koch, A.E. Angiogenesis in Rheumatoid Arthritis. Autoimmunity 2009, 42, 563–573. [Google Scholar] [CrossRef] [Green Version]
- Pelechas, E.; Voulgari, P.V.; Drosos, A.A. Preclinical Discovery and Development of Adalimumab for the Treatment of Rheumatoid Arthritis. Expert Opin. Drug Discov. 2021, 16, 227–234. [Google Scholar] [CrossRef] [PubMed]
- Smith, G.P. Filamentous Fusion Phage: Novel Expression Vectors That Display Cloned Antigens on the Virion Surface. Science 1985, 228, 1315–1317. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Chadwick, L.; Mysler, E.; Moots, R.J. Review of Biosimilar Trials and Data on Adalimumab in Rheumatoid Arthritis. Curr. Rheumatol. Rep. 2018, 20, 57. [Google Scholar] [CrossRef] [PubMed]
- Cvetković, R.S.; Scott, L.J. Adalimumab. BioDrugs 2006, 20, 293–311. [Google Scholar] [CrossRef] [PubMed]
- Avila-Pedretti, G.; Tornero, J.; Fernández-Nebro, A.; Blanco, F.; González-Alvaro, I.; Cañete, J.D.; Maymó, J.; Alperiz, M.; Fernández-Gutiérrez, B.; Olivé, A.; et al. Variation at FCGR2A and Functionally Related Genes Is Associated with the Response to Anti-TNF Therapy in Rheumatoid Arthritis. PLoS ONE 2015, 10, e0122088. [Google Scholar] [CrossRef] [Green Version]
- Dávila-Fajardo, C.L.; Márquez, A.; Pascual-Salcedo, D.; Moreno Ramos, M.J.; García-Portales, R.; Magro, C.; Alegre-Sancho, J.J.; Balsa, A.; Cabeza-Barrera, J.; Raya, E.; et al. Confirmation of -174G/C Interleukin-6 Gene Promoter Polymorphism as a Genetic Marker Predicting Antitumor Necrosis Factor Treatment Outcome. Pharm. Genom. 2014, 24, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Potter, C.; Hyrich, K.L.; Tracey, A.; Lunt, M.; Plant, D.; Symmons, D.P.M.; Thomson, W.; Worthington, J.; Emery, P.; Morgan, A.W.; et al. Association of Rheumatoid Factor and Anti-Cyclic Citrullinated Peptide Positivity, but Not Carriage of Shared Epitope or PTPN22 Susceptibility Variants, with Anti-Tumour Necrosis Factor Response in Rheumatoid Arthritis. Ann. Rheum. Dis. 2009, 68, 69–74. [Google Scholar] [CrossRef] [Green Version]
- Cuchacovich, M.; Soto, L.; Edwardes, M.; Gutierrez, M.; Llanos, C.; Pacheco, D.; Sabugo, F.; Alamo, M.; Fuentealba, C.; Villanueva, L.; et al. Tumour Necrosis Factor (TNF)Alpha -308 G/G Promoter Polymorphism and TNFalpha Levels Correlate with a Better Response to Adalimumab in Patients with Rheumatoid Arthritis. Scand. J. Rheumatol. 2006, 35, 435–440. [Google Scholar] [CrossRef]
- Seitz, M.; Wirthmüller, U.; Möller, B.; Villiger, P.M. The -308 Tumour Necrosis Factor-Alpha Gene Polymorphism Predicts Therapeutic Response to TNFalpha-Blockers in Rheumatoid Arthritis and Spondyloarthritis Patients. Rheumatology 2007, 46, 93–96. [Google Scholar] [CrossRef] [Green Version]
- O’Rielly, D.D.; Roslin, N.M.; Beyene, J.; Pope, A.; Rahman, P. TNF-Alpha-308 G/A Polymorphism and Responsiveness to TNF-Alpha Blockade Therapy in Moderate to Severe Rheumatoid Arthritis: A Systematic Review and Meta-Analysis. Pharm. J. 2009, 9, 161–167. [Google Scholar] [CrossRef]
- Zeng, Z.; Duan, Z.; Zhang, T.; Wang, S.; Li, G.; Gao, J.; Ye, D.; Xu, S.; Xu, J.; Zhang, L.; et al. Association between Tumor Necrosis Factor-α (TNF-α) Promoter -308 G/A and Response to TNF-α Blockers in Rheumatoid Arthritis: A Meta-Analysis. Mod. Rheumatol. 2013, 23, 489–495. [Google Scholar] [CrossRef]
- Miceli-Richard, C.; Comets, E.; Verstuyft, C.; Tamouza, R.; Loiseau, P.; Ravaud, P.; Kupper, H.; Becquemont, L.; Charron, D.; Mariette, X. A Single Tumour Necrosis Factor Haplotype Influences the Response to Adalimumab in Rheumatoid Arthritis. Ann. Rheum. Dis. 2008, 67, 478–484. [Google Scholar] [CrossRef]
- Ongaro, A.; De Mattei, M.; Pellati, A.; Caruso, A.; Ferretti, S.; Masieri, F.F.; Fotinidi, M.; Farina, I.; Trotta, F.; Padovan, M. Can Tumor Necrosis Factor Receptor II Gene 676T>G Polymorphism Predict the Response Grading to Anti-TNFalpha Therapy in Rheumatoid Arthritis? Rheumatol. Int. 2008, 28, 901–908. [Google Scholar] [CrossRef]
- Sibéril, S.; Dutertre, C.-A.; Boix, C.; Bonnin, E.; Ménez, R.; Stura, E.; Jorieux, S.; Fridman, W.-H.; Teillaud, J.-L. Molecular Aspects of Human FcgammaR Interactions with IgG: Functional and Therapeutic Consequences. Immunol. Lett. 2006, 106, 111–118. [Google Scholar] [CrossRef]
- Musolino, A.; Naldi, N.; Bortesi, B.; Pezzuolo, D.; Capelletti, M.; Missale, G.; Laccabue, D.; Zerbini, A.; Camisa, R.; Bisagni, G.; et al. Immunoglobulin G Fragment C Receptor Polymorphisms and Clinical Efficacy of Trastuzumab-Based Therapy in Patients with HER-2/Neu-Positive Metastatic Breast Cancer. J. Clin. Oncol. 2008, 26, 1789–1796. [Google Scholar] [CrossRef]
- Guilliams, M.; Bruhns, P.; Saeys, Y.; Hammad, H.; Lambrecht, B.N. The Function of Fcγ Receptors in Dendritic Cells and Macrophages. Nat. Rev. Immunol. 2014, 14, 94–108. [Google Scholar] [CrossRef] [PubMed]
- Montes, A.; Perez-Pampin, E.; Narváez, J.; Cañete, J.D.; Navarro-Sarabia, F.; Moreira, V.; Fernández-Nebro, A.; Del Carmen Ordóñez, M.; de la Serna, A.R.; Magallares, B.; et al. Association of FCGR2A with the Response to Infliximab Treatment of Patients with Rheumatoid Arthritis. Pharm. Genom. 2014, 24, 238–245. [Google Scholar] [CrossRef] [PubMed]
- Arora, T.; Padaki, R.; Liu, L.; Hamburger, A.E.; Ellison, A.R.; Stevens, S.R.; Louie, J.S.; Kohno, T. Differences in Binding and Effector Functions between Classes of TNF Antagonists. Cytokine 2009, 45, 124–131. [Google Scholar] [CrossRef] [PubMed]
- Kurreeman, F.; Liao, K.; Chibnik, L.; Hickey, B.; Stahl, E.; Gainer, V.; Li, G.; Bry, L.; Mahan, S.; Ardlie, K.; et al. Genetic Basis of Autoantibody Positive and Negative Rheumatoid Arthritis Risk in a Multi-Ethnic Cohort Derived from Electronic Health Records. Am. J. Hum. Genet. 2011, 88, 57–69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bossini-Castillo, L.; de Kovel, C.; Kallberg, H.; van ’t Slot, R.; Italiaander, A.; Coenen, M.; Tak, P.P.; Posthumus, M.D.; Wijmenga, C.; Huizinga, T.; et al. A Genome-Wide Association Study of Rheumatoid Arthritis without Antibodies against Citrullinated Peptides. Ann. Rheum. Dis. 2015, 74, e15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdelhaleem, M. The Novel Helicase Homologue DDX32 Is Down-Regulated in Acute Lymphoblastic Leukemia. Leuk. Res. 2002, 26, 945–954. [Google Scholar] [CrossRef]
- Abdelhaleem, M.; Sun, T.-H.; Ho, M. DHX32 Expression Suggests a Role in Lymphocyte Differentiation. Anticancer Res. 2005, 25, 2645–2648. [Google Scholar]
- Meylan, E.; Tschopp, J. Toll-like Receptors and RNA Helicases: Two Parallel Ways to Trigger Antiviral Responses. Mol. Cell 2006, 22, 561–569. [Google Scholar] [CrossRef]
- Baccala, R.; Gonzalez-Quintial, R.; Lawson, B.R.; Stern, M.E.; Kono, D.H.; Beutler, B.; Theofilopoulos, A.N. Sensors of the Innate Immune System: Their Mode of Action. Nat. Rev. Rheumatol. 2009, 5, 448–456. [Google Scholar] [CrossRef]
- Yuan, G.; Yang, S.; Ng, A.; Fu, C.; Oursler, M.J.; Xing, L.; Yang, S. RGS12 Is a Novel Critical NF-ΚB Activator in Inflammatory Arthritis. iScience 2020, 23, 101172. [Google Scholar] [CrossRef]
- Yuan, G.; Yang, S.; Gautam, M.; Luo, W.; Yang, S. Macrophage Regulator of G-Protein Signaling 12 Contributes to Inflammatory Pain Hypersensitivity. Ann. Transl. Med. 2021, 9, 448. [Google Scholar] [CrossRef]
- Mugnier, B.; Balandraud, N.; Darque, A.; Roudier, C.; Roudier, J.; Reviron, D. Polymorphism at Position -308 of the Tumor Necrosis Factor Alpha Gene Influences Outcome of Infliximab Therapy in Rheumatoid Arthritis. Arthritis Rheum. 2003, 48, 1849–1852. [Google Scholar] [CrossRef]
- Padyukov, L.; Lampa, J.; Heimbürger, M.; Ernestam, S.; Cederholm, T.; Lundkvist, I.; Andersson, P.; Hermansson, Y.; Harju, A.; Klareskog, L.; et al. Genetic Markers for the Efficacy of Tumour Necrosis Factor Blocking Therapy in Rheumatoid Arthritis. Ann. Rheum. Dis. 2003, 62, 526–529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cuchacovich, M.; Ferreira, L.; Aliste, M.; Soto, L.; Cuenca, J.; Cruzat, A.; Gatica, H.; Schiattino, I.; Pérez, C.; Aguirre, A.; et al. Tumour Necrosis Factor-Alpha (TNF-Alpha) Levels and Influence of -308 TNF-Alpha Promoter Polymorphism on the Responsiveness to Infliximab in Patients with Rheumatoid Arthritis. Scand. J. Rheumatol. 2004, 33, 228–232. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, J.E.; Carvalho, T.; Cruz, M.; Nero, P.; Sobral, M.; Mourão, A.F.; Cavaleiro, J.; Ligeiro, D.; Abreu, I.; Carmo-Fonseca, M.; et al. Polymorphism at Position -308 of the Tumour Necrosis Factor Alpha Gene and Rheumatoid Arthritis Pharmacogenetics. Ann. Rheum. Dis. 2005, 64, 793–794. [Google Scholar] [CrossRef] [Green Version]
- Kang, C.P.; Lee, K.W.; Yoo, D.H.; Kang, C.; Bae, S.C. The Influence of a Polymorphism at Position -857 of the Tumour Necrosis Factor Alpha Gene on Clinical Response to Etanercept Therapy in Rheumatoid Arthritis. Rheumatology 2005, 44, 547–552. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guis, S.; Balandraud, N.; Bouvenot, J.; Auger, I.; Toussirot, E.; Wendling, D.; Mattei, J.-P.; Nogueira, L.; Mugnier, B.; Legeron, P.; et al. Influence of -308 A/G Polymorphism in the Tumor Necrosis Factor Alpha Gene on Etanercept Treatment in Rheumatoid Arthritis. Arthritis Rheum. 2007, 57, 1426–1430. [Google Scholar] [CrossRef] [PubMed]
- Marotte, H.; Arnaud, B.; Diasparra, J.; Zrioual, S.; Miossec, P. Association between the Level of Circulating Bioactive Tumor Necrosis Factor Alpha and the Tumor Necrosis Factor Alpha Gene Polymorphism at -308 in Patients with Rheumatoid Arthritis Treated with a Tumor Necrosis Factor Alpha Inhibitor. Arthritis Rheum. 2008, 58, 1258–1263. [Google Scholar] [CrossRef]
- Balog, A.; Gál, J.; Gyulai, Z.; Zsilák, S.; Mándi, Y. Tumour Necrosis Factor-Alpha and Heat-Shock Protein 70-2 Gene Polymorphisms in a Family with Rheumatoid Arthritis. Acta Microbiol. Immunol. Hung. 2004, 51, 263–269. [Google Scholar] [CrossRef]
- Criswell, L.A.; Lum, R.F.; Turner, K.N.; Woehl, B.; Zhu, Y.; Wang, J.; Tiwari, H.K.; Edberg, J.C.; Kimberly, R.P.; Moreland, L.W.; et al. The Influence of Genetic Variation in the HLA-DRB1 and LTA-TNF Regions on the Response to Treatment of Early Rheumatoid Arthritis with Methotrexate or Etanercept. Arthritis Rheum. 2004, 50, 2750–2756. [Google Scholar] [CrossRef]
- Chatzikyriakidou, A.; Georgiou, I.; Voulgari, P.V.; Venetsanopoulou, A.I.; Drosos, A.A. Combined Tumour Necrosis Factor-Alpha and Tumour Necrosis Factor Receptor Genotypes Could Predict Rheumatoid Arthritis Patients’ Response to Anti-TNF-Alpha Therapy and Explain Controversies of Studies Based on a Single Polymorphism. Rheumatology 2007, 46, 1034–1035. [Google Scholar] [CrossRef] [Green Version]
- Maxwell, J.R.; Potter, C.; Hyrich, K.L.; Biologics in Rheumatoid Arthritis Genetics and Genomics Study Syndicate; Barton, A.; Worthington, J.; Isaacs, J.D.; Morgan, A.W.; Wilson, A.G. Association of the Tumour Necrosis Factor-308 Variant with Differential Response to Anti-TNF Agents in the Treatment of Rheumatoid Arthritis. Hum. Mol. Genet. 2008, 17, 3532–3538. [Google Scholar] [CrossRef] [Green Version]
- Pavy, S.; Toonen, E.J.M.; Miceli-Richard, C.; Barrera, P.; van Riel, P.L.C.M.; Criswell, L.A.; Mariette, X.; Coenen, M.J.H. Tumour Necrosis Factor Alpha -308G->A Polymorphism Is Not Associated with Response to TNFalpha Blockers in Caucasian Patients with Rheumatoid Arthritis: Systematic Review and Meta-Analysis. Ann. Rheum. Dis. 2010, 69, 1022–1028. [Google Scholar] [CrossRef]
- Felson, D.T.; Anderson, J.J.; Boers, M.; Bombardier, C.; Furst, D.; Goldsmith, C.; Katz, L.M.; Lightfoot, R.; Paulus, H.; Strand, V. American College of Rheumatology. Preliminary Definition of Improvement in Rheumatoid Arthritis. Arthritis Rheum. 1995, 38, 727–735. [Google Scholar] [CrossRef]
- Lichtenstein, L.; Ron, Y.; Kivity, S.; Ben-Horin, S.; Israeli, E.; Fraser, G.M.; Dotan, I.; Chowers, Y.; Confino-Cohen, R.; Weiss, B. Infliximab-Related Infusion Reactions: Systematic Review. J. Crohns Colitis 2015, 9, 806–815. [Google Scholar] [CrossRef] [Green Version]
- Hanauer, S.B.; Feagan, B.G.; Lichtenstein, G.R.; Mayer, L.F.; Schreiber, S.; Colombel, J.F.; Rachmilewitz, D.; Wolf, D.C.; Olson, A.; Bao, W.; et al. Maintenance Infliximab for Crohn’s Disease: The ACCENT I Randomised Trial. Lancet 2002, 359, 1541–1549. [Google Scholar] [CrossRef]
- Rutgeerts, P.; Sandborn, W.J.; Feagan, B.G.; Reinisch, W.; Olson, A.; Johanns, J.; Travers, S.; Rachmilewitz, D.; Hanauer, S.B.; Lichtenstein, G.R.; et al. Infliximab for Induction and Maintenance Therapy for Ulcerative Colitis. N. Engl. J. Med. 2005, 353, 2462–2476. [Google Scholar] [CrossRef] [Green Version]
- Markham, A.; Lamb, H.M. Infliximab. Drugs 2000, 59, 1341–1359. [Google Scholar] [CrossRef]
- Cañete, J.D.; Suárez, B.; Hernández, M.V.; Sanmartí, R.; Rego, I.; Celis, R.; Moll, C.; Pinto, J.A.; Blanco, F.J.; Lozano, F. Influence of Variants of Fc Gamma Receptors IIA and IIIA on the American College of Rheumatology and European League Against Rheumatism Responses to Anti-Tumour Necrosis Factor Alpha Therapy in Rheumatoid Arthritis. Ann. Rheum. Dis. 2009, 68, 1547–1552. [Google Scholar] [CrossRef]
- Martinez, A.; Salido, M.; Bonilla, G.; Pascual-Salcedo, D.; Fernandez-Arquero, M.; de Miguel, S.; Balsa, A.; de la Concha, E.G.; Fernandez-Gutierrez, B. Association of the Major Histocompatibility Complex with Response to Infliximab Therapy in Rheumatoid Arthritis Patients. Arthritis Rheum. 2004, 50, 1077–1082. [Google Scholar] [CrossRef]
- Fabris, M.; Di Poi, E.; D’Elia, A.; Damante, G.; Sinigaglia, L.; Ferraccioli, G. Tumor Necrosis Factor-Alpha Gene Polymorphism in Severe and Mild-Moderate Rheumatoid Arthritis. J. Rheumatol. 2002, 29, 29–33. [Google Scholar] [PubMed]
- Swierkot, J.; Bogunia-Kubik, K.; Nowak, B.; Bialowas, K.; Korman, L.; Gebura, K.; Kolossa, K.; Jeka, S.; Wiland, P. Analysis of Associations between Polymorphisms within Genes Coding for Tumour Necrosis Factor (TNF)-Alpha and TNF Receptors and Responsiveness to TNF-Alpha Blockers in Patients with Rheumatoid Arthritis. Jt. Bone Spine 2015, 82, 94–99. [Google Scholar] [CrossRef]
- van Sorge, N.M.; van der Pol, W.-L.; van de Winkel, J.G.J. FcgammaR Polymorphisms: Implications for Function, Disease Susceptibility and Immunotherapy. Tissue Antigens 2003, 61, 189–202. [Google Scholar] [CrossRef] [PubMed]
- Barrera, P.; Joosten, L.A.; den Broeder, A.A.; van de Putte, L.B.; van Riel, P.L.; van den Berg, W.B. Effects of Treatment with a Fully Human Anti-Tumour Necrosis Factor Alpha Monoclonal Antibody on the Local and Systemic Homeostasis of Interleukin 1 and TNFalpha in Patients with Rheumatoid Arthritis. Ann. Rheum. Dis. 2001, 60, 660–669. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edwards, J.C.; Cambridge, G.; Abrahams, V.M. Do Self-Perpetuating B Lymphocytes Drive Human Autoimmune Disease? Immunology 1999, 97, 188–196. [Google Scholar] [CrossRef]
- Dörner, T.; Jacobi, A.M.; Lipsky, P.E. B Cells in Autoimmunity. Arthritis Res. Ther. 2009, 11, 247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McInnes, I.B.; Schett, G. The Pathogenesis of Rheumatoid Arthritis. N. Engl. J. Med. 2011, 365, 2205–2219. [Google Scholar] [CrossRef] [Green Version]
- Mota, P.; Reddy, V.; Isenberg, D. Improving B-Cell Depletion in Systemic Lupus Erythematosus and Rheumatoid Arthritis. Expert Rev. Clin. Immunol. 2017, 13, 667–676. [Google Scholar] [CrossRef]
- Salles, G.; Barrett, M.; Foà, R.; Maurer, J.; O’Brien, S.; Valente, N.; Wenger, M.; Maloney, D.G. Rituximab in B-Cell Hematologic Malignancies: A Review of 20 Years of Clinical Experience. Adv. Ther. 2017, 34, 2232–2273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- García-Muñoz, R.; Quero, C.; Pérez-Persona, E.; Domingo-García, A.; Pérez-López, C.; Villaescusa-de-la-Rosa, T.; Martínez-Castro, A.M.; Arguiñano-Pérez, J.M.; Parra-Cuadrado, J.F.; Panizo, C. Safety of Switching from Intravenous to Subcutaneous Rituximab during First-Line Treatment of Patients with Non-Hodgkin Lymphoma: The Spanish Population of the MabRella Study. Br. J. Haematol. 2020, 188, 661–673. [Google Scholar] [CrossRef] [Green Version]
- Cylwik, B.; Gruszewska, E.; Gindzienska-Sieskiewicz, E.; Kowal-Bielecka, O.; Chrostek, L. Serum Profile of Transferrin Isoforms in Rheumatoid Arthritis Treated with Biological Drugs. Clin. Biochem. 2019, 74, 31–35. [Google Scholar] [CrossRef]
- von Borstel, A.; Land, J.; Abdulahad, W.H.; Rutgers, A.; Stegeman, C.A.; Diepstra, A.; Heeringa, P.; Sanders, J.S. CD27+CD38hi B Cell Frequency During Remission Predicts Relapsing Disease in Granulomatosis With Polyangiitis Patients. Front. Immunol. 2019, 10, 2221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Golay, J.; Semenzato, G.; Rambaldi, A.; Foà, R.; Gaidano, G.; Gamba, E.; Pane, F.; Pinto, A.; Specchia, G.; Zaja, F.; et al. Lessons for the Clinic from Rituximab Pharmacokinetics and Pharmacodynamics. mAbs 2013, 5, 826–837. [Google Scholar] [CrossRef] [PubMed]
- Weiner, G.J. Rituximab: Mechanism of Action. Semin. Hematol. 2010, 47, 115–123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uchida, J.; Hamaguchi, Y.; Oliver, J.A.; Ravetch, J.V.; Poe, J.C.; Haas, K.M.; Tedder, T.F. The Innate Mononuclear Phagocyte Network Depletes B Lymphocytes through Fc Receptor-Dependent Mechanisms during Anti-CD20 Antibody Immunotherapy. J. Exp. Med. 2004, 199, 1659–1669. [Google Scholar] [CrossRef] [PubMed]
- Bergantini, L.; d’Alessandro, M.; Cameli, P.; Vietri, L.; Vagaggini, C.; Perrone, A.; Sestini, P.; Frediani, B.; Bargagli, E. Effects of Rituximab Therapy on B Cell Differentiation and Depletion. Clin. Rheumatol. 2020, 39, 1415–1421. [Google Scholar] [CrossRef] [PubMed]
- Jiménez Morales, A.; Maldonado-Montoro, M.; Martínez de la Plata, J.E.; Pérez Ramírez, C.; Daddaoua, A.; Alarcón Payer, C.; Expósito Ruiz, M.; García Collado, C. FCGR2A/FCGR3A Gene Polymorphisms and Clinical Variables as Predictors of Response to Tocilizumab and Rituximab in Patients With Rheumatoid Arthritis. J. Clin. Pharmacol. 2019, 59, 517–531. [Google Scholar] [CrossRef]
- Quartuccio, L.; Fabris, M.; Pontarini, E.; Salvin, S.; Zabotti, A.; Benucci, M.; Manfredi, M.; Biasi, D.; Ravagnani, V.; Atzeni, F.; et al. The 158VV Fcgamma Receptor 3A Genotype Is Associated with Response to Rituximab in Rheumatoid Arthritis: Results of an Italian Multicentre Study. Ann. Rheum. Dis. 2014, 73, 716–721. [Google Scholar] [CrossRef] [PubMed]
- Ruyssen-Witrand, A.; Rouanet, S.; Combe, B.; Dougados, M.; Le Loët, X.; Sibilia, J.; Tebib, J.; Mariette, X.; Constantin, A. Fcγ Receptor Type IIIA Polymorphism Influences Treatment Outcomes in Patients with Rheumatoid Arthritis Treated with Rituximab. Ann. Rheum. Dis. 2012, 71, 875–877. [Google Scholar] [CrossRef]
- Pál, I.; Szamosi, S.; Hodosi, K.; Szekanecz, Z.; Váróczy, L. Effect of Fcγ-Receptor 3a (FCGR3A) Gene Polymorphisms on Rituximab Therapy in Hungarian Patients with Rheumatoid Arthritis. RMD Open 2017, 3, e000485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sarsour, K.; Greenberg, J.; Johnston, J.A.; Nelson, D.R.; O’Brien, L.A.; Oddoux, C.; Ostrer, H.; Pearlman, A.; Reed, G. The Role of the FcGRIIIa Polymorphism in Modifying the Association between Treatment and Outcome in Patients with Rheumatoid Arthritis Treated with Rituximab versus TNF-α Antagonist Therapies. Clin. Exp. Rheumatol. 2013, 31, 189–194. [Google Scholar]
- Ruyssen-Witrand, A.; Rouanet, S.; Combe, B.; Dougados, M.; Le Loët, X.; Sibilia, J.; Tebib, J.; Mariette, X.; Constantin, A. Association between -871C>T Promoter Polymorphism in the B-Cell Activating Factor Gene and the Response to Rituximab in Rheumatoid Arthritis Patients. Rheumatology 2013, 52, 636–641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fabris, M.; Quartuccio, L.; Lombardi, S.; Benucci, M.; Manfredi, M.; Saracco, M.; Atzeni, F.; Morassi, P.; Cimmino, M.A.; Pontarini, E.; et al. Study on the Possible Role of the -174G>C IL-6 Promoter Polymorphism in Predicting Response to Rituximab in Rheumatoid Arthritis. Reumatismo 2010, 62, 253–258. [Google Scholar] [CrossRef] [PubMed]
- Daïen, C.I.; Fabre, S.; Rittore, C.; Soler, S.; Daïen, V.; Tejedor, G.; Cadart, D.; Molinari, N.; Daurès, J.-P.; Jorgensen, C.; et al. TGF Beta1 Polymorphisms Are Candidate Predictors of the Clinical Response to Rituximab in Rheumatoid Arthritis. Jt. Bone Spine 2012, 79, 471–475. [Google Scholar] [CrossRef] [PubMed]
- Chihara, N.; Aranami, T.; Sato, W.; Miyazaki, Y.; Miyake, S.; Okamoto, T.; Ogawa, M.; Toda, T.; Yamamura, T. Interleukin 6 Signaling Promotes Anti-Aquaporin 4 Autoantibody Production from Plasmablasts in Neuromyelitis Optica. Proc. Natl. Acad. Sci. USA 2011, 108, 3701–3706. [Google Scholar] [CrossRef] [Green Version]
- Narazaki, M.; Tanaka, T.; Kishimoto, T. The Role and Therapeutic Targeting of IL-6 in Rheumatoid Arthritis. Expert Rev. Clin. Immunol. 2017, 13, 535–551. [Google Scholar] [CrossRef]
- Mihara, M.; Kasutani, K.; Okazaki, M.; Nakamura, A.; Kawai, S.; Sugimoto, M.; Matsumoto, Y.; Ohsugi, Y. Tocilizumab Inhibits Signal Transduction Mediated by Both MIL-6R and SIL-6R, but Not by the Receptors of Other Members of IL-6 Cytokine Family. Int. Immunopharmacol. 2005, 5, 1731–1740. [Google Scholar] [CrossRef]
- Snir, A.; Kessel, A.; Haj, T.; Rosner, I.; Slobodin, G.; Toubi, E. Anti-IL-6 Receptor Antibody (Tocilizumab): A B Cell Targeting Therapy. Clin. Exp. Rheumatol. 2011, 29, 697–700. [Google Scholar] [CrossRef] [Green Version]
- Scott, L.J. Tocilizumab: A Review in Rheumatoid Arthritis. Drugs 2017, 77, 1865–1879. [Google Scholar] [CrossRef] [Green Version]
- Dhillon, S. Intravenous Tocilizumab: A Review of Its Use in Adults with Rheumatoid Arthritis. BioDrugs Clin. Immunother. Biopharm. Gene Ther. 2014, 28, 75–106. [Google Scholar] [CrossRef]
- Wang, J.; Bansal, A.T.; Martin, M.; Germer, S.; Benayed, R.; Essioux, L.; Lee, J.S.; Begovich, A.; Hemmings, A.; Kenwright, A.; et al. Genome-Wide Association Analysis Implicates the Involvement of Eight Loci with Response to Tocilizumab for the Treatment of Rheumatoid Arthritis. Pharm. J. 2013, 13, 235–241. [Google Scholar] [CrossRef] [Green Version]
- Maldonado-Montoro, M.; Cañadas-Garre, M.; González-Utrilla, A.; Ángel Calleja-Hernández, M. Influence of IL6R Gene Polymorphisms in the Effectiveness to Treatment with Tocilizumab in Rheumatoid Arthritis. Pharm. J. 2018, 18, 167–172. [Google Scholar] [CrossRef] [PubMed]
- Gregersen, P.K.; Silver, J.; Winchester, R.J. The Shared Epitope Hypothesis. An Approach to Understanding the Molecular Genetics of Susceptibility to Rheumatoid Arthritis. Arthritis Rheum. 1987, 30, 1205–1213. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Wang, Y.; Xu, D.; Nossent, J.; Pavlos, N.J.; Xu, J. Rheumatoid Arthritis: Pathological Mechanisms and Modern Pharmacologic Therapies. Bone Res. 2018, 6, 15. [Google Scholar] [CrossRef]
- Pallio, G.; Mannino, F.; Irrera, N.; Eid, A.H.; Squadrito, F.; Bitto, A. Polymorphisms Involved in Response to Biological Agents Used in Rheumatoid Arthritis. Biomolecules 2020, 10, 1203. [Google Scholar] [CrossRef]
- Kaijzel, E.L.; van Dongen, H.; Bakker, A.M.; Breedveld, F.C.; Huizinga, T.W.J.; Verweij, C.L. Relationship of Polymorphisms of the Interleukin-1 Gene Cluster to Occurrence and Severity of Rheumatoid Arthritis. Tissue Antigens 2002, 59, 122–126. [Google Scholar] [CrossRef] [PubMed]
- Camargo, J.F.; Correa, P.A.; Castiblanco, J.; Anaya, J.-M. Interleukin-1beta Polymorphisms in Colombian Patients with Autoimmune Rheumatic Diseases. Genes Immun. 2004, 5, 609–614. [Google Scholar] [CrossRef] [PubMed]
- Buchs, N.; di Giovine, F.S.; Silvestri, T.; Vannier, E.; Duff, G.W.; Miossec, P. IL-1B and IL-1Ra Gene Polymorphisms and Disease Severity in Rheumatoid Arthritis: Interaction with Their Plasma Levels. Genes Immun. 2001, 2, 222–228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arman, A.; Yilmaz, B.; Coker, A.; Inanc, N.; Direskeneli, H. Interleukin-1 Receptor Antagonist (IL-1RN) and Interleukin-1B Gene Polymorphisms in Turkish Patients with Rheumatoid Arthritis. Clin. Exp. Rheumatol. 2006, 24, 643–648. [Google Scholar]
- Tolusso, B.; Pietrapertosa, D.; Morelli, A.; De Santis, M.; Gremese, E.; Farina, G.; Carniello, S.G.; Del Frate, M.; Ferraccioli, G. IL-1B and IL-1RN Gene Polymorphisms in Rheumatoid Arthritis: Relationship with Protein Plasma Levels and Response to Therapy. Pharmacogenomics 2006, 7, 683–695. [Google Scholar] [CrossRef]
- Nordang, G.B.N.; Viken, M.K.; Hollis-Moffatt, J.E.; Merriman, T.R.; Førre, Ø.T.; Helgetveit, K.; Kvien, T.K.; Lie, B.A. Association Analysis of the Interleukin 17A Gene in Caucasian Rheumatoid Arthritis Patients from Norway and New Zealand. Rheumatology 2009, 48, 367–370. [Google Scholar] [CrossRef] [Green Version]
- Paradowska-Gorycka, A.; Wojtecka-Lukasik, E.; Trefler, J.; Wojciechowska, B.; Lacki, J.K.; Maslinski, S. Association between IL-17F Gene Polymorphisms and Susceptibility to and Severity of Rheumatoid Arthritis (RA). Scand. J. Immunol. 2010, 72, 134–141. [Google Scholar] [CrossRef]
- Han, L.; Lee, H.S.; Yoon, J.H.; Choi, W.S.; Park, Y.G.; Nam, S.W.; Lee, J.Y.; Park, W.S. Association of IL-17A and IL-17F Single Nucleotide Polymorphisms with Susceptibility to Osteoarthritis in a Korean Population. Gene 2014, 533, 119–122. [Google Scholar] [CrossRef] [PubMed]
- Erkol İnal, E.; Görükmez, O.; Dündar, Ü.; Görükmez, Ö.; Yener, M.; Özemri Sağ, Ş.; Yakut, T. The Influence of Polymorphisms of Interleukin-17A and -17F Genes on Susceptibility and Activity of Rheumatoid Arthritis. Genet. Test. Mol. Biomark. 2015, 19, 461–464. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Zhang, H.; Yan, T.; Zhou, G.; Liu, R. Association between Interleukin 17A Polymorphisms and Susceptibility to Rheumatoid Arthritis in a Chinese Population. Gene 2015, 566, 18–22. [Google Scholar] [CrossRef] [PubMed]
- Bogunia-Kubik, K.; Świerkot, J.; Malak, A.; Wysoczańska, B.; Nowak, B.; Białowąs, K.; Gębura, K.; Korman, L.; Wiland, P. IL-17A, IL-17F and IL-23R Gene Polymorphisms in Polish Patients with Rheumatoid Arthritis. Arch. Immunol. Ther. Exp. 2015, 63, 215–221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carvalho, C.N.; do Carmo, R.F.; Duarte, A.L.P.; Carvalho, A.A.T.; Leão, J.C.; Gueiros, L.A. IL-17A and IL-17F Polymorphisms in Rheumatoid Arthritis and Sjögren’s Syndrome. Clin. Oral Investig. 2016, 20, 495–502. [Google Scholar] [CrossRef] [PubMed]
- Pawlik, A.; Kotrych, D.; Malinowski, D.; Dziedziejko, V.; Czerewaty, M.; Safranow, K. IL17A and IL17F Gene Polymorphisms in Patients with Rheumatoid Arthritis. BMC Musculoskelet. Disord. 2016, 17, 208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Louahchi, S.; Allam, I.; Berkani, L.; Boucharef, A.; Abdesemed, A.; Khaldoun, N.; Nebbab, A.; Ladjouze, A.; Djidjik, R. Association Study of Single Nucleotide Polymorphisms of IL23R and IL17 in Rheumatoid Arthritis in the Algerian Population. Acta Reumatol. Port. 2016, 41, 151–157. [Google Scholar] [PubMed]
- Marwa, O.S.; Kalthoum, T.; Wajih, K.; Kamel, H. Association of IL17A and IL17F Genes with Rheumatoid Arthritis Disease and the Impact of Genetic Polymorphisms on Response to Treatment. Immunol. Lett. 2017, 183, 24–36. [Google Scholar] [CrossRef]
- Gomes da Silva, I.I.F.; Angelo, H.D.; Rushansky, E.; Mariano, M.H.; de Mascena Diniz Maia, M.; de Souza, P.R.E. Interleukin (IL)-23 Receptor, IL-17A and IL-17F Gene Polymorphisms in Brazilian Patients with Rheumatoid Arthritis. Arch. Immunol. Ther. Exp. 2017, 65, 537–543. [Google Scholar] [CrossRef]
- de la Peña, M.G.; Cruz, R.M.; Guerrero, E.G.; López, A.G.; Molina, G.P.; González, N.E.H. Polymorphism Rs2275913 of Interleukin-17A Is Related to More Intensive Therapy with Disease-Modifying Anti Rheumatic Drugs in Mexican Patients with Rheumatoid Arthritis. Acta Reumatol. Port. 2017, 42, 155–161. [Google Scholar]
- Lee, Y.H.; Bae, S.-C. Associations between Circulating IL-17 Levels and Rheumatoid Arthritis and between IL-17 Gene Polymorphisms and Disease Susceptibility: A Meta-Analysis. Postgrad. Med. J. 2017, 93, 465–471. [Google Scholar] [CrossRef] [PubMed]
- Dhaouadi, T.; Chahbi, M.; Haouami, Y.; Sfar, I.; Abdelmoula, L.; Ben Abdallah, T.; Gorgi, Y. IL-17A, IL-17RC Polymorphisms and IL17 Plasma Levels in Tunisian Patients with Rheumatoid Arthritis. PLoS ONE 2018, 13, e0194883. [Google Scholar] [CrossRef] [PubMed]
- Agonia, I.; Couras, J.; Cunha, A.; Andrade, A.J.; Macedo, J.; Sousa-Pinto, B. IL-17, IL-21 and IL-22 Polymorphisms in Rheumatoid Arthritis: A Systematic Review and Meta-Analysis. Cytokine 2020, 125, 154813. [Google Scholar] [CrossRef]
- Nisar, H.; Pasha, U.; Mirza, M.U.; Abid, R.; Hanif, K.; Kadarmideen, H.N.; Sadaf, S. Impact of IL-17F 7488T/C Functional Polymorphism on Progressive Rheumatoid Arthritis: Novel Insight from the Molecular Dynamic Simulations. Immunol. Investig. 2021, 50, 416–426. [Google Scholar] [CrossRef]
- Amin, A.; Sheikh, N.; Mukhtar, M.; Saleem, T.; Akhtar, T.; Fatima, N.; Mehmood, R. Association of Interleukin-17 Gene Polymorphisms with the Onset of Rheumatoid Arthritis. Immunobiology 2021, 226, 152045. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Li, Y.; Li, L.; Zhang, G.; Zhang, F.; Tang, Y.; Zhou, L.; Yang, Y.; Li, J. Association between the Interleukin (IL)-17A Rs2275913 Polymorphism and Rheumatoid Arthritis Susceptibility: A Meta-Analysis and Trial Sequential Analysis. J. Int. Med. Res. 2021, 49, 3000605211053233. [Google Scholar] [CrossRef] [PubMed]
- Shao, M.; Xu, S.; Yang, H.; Xu, W.; Deng, J.; Chen, Y.; Gao, X.; Guan, S.; Xu, S.; Shuai, Z.; et al. Association between IL-17A and IL-17F Gene Polymorphism and Susceptibility in Inflammatory Arthritis: A Meta-Analysis. Clin. Immunol. 2020, 213, 108374. [Google Scholar] [CrossRef]
- Orozco, G.; Rueda, B.; Robledo, G.; García, A.; Martín, J. Investigation of the IL23R Gene in a Spanish Rheumatoid Arthritis Cohort. Hum. Immunol. 2007, 68, 681–684. [Google Scholar] [CrossRef] [PubMed]
- Faragó, B.; Magyari, L.; Sáfrány, E.; Csöngei, V.; Járomi, L.; Horvatovich, K.; Sipeky, C.; Maász, A.; Radics, J.; Gyetvai, A.; et al. Functional Variants of Interleukin-23 Receptor Gene Confer Risk for Rheumatoid Arthritis but Not for Systemic Sclerosis. Ann. Rheum. Dis. 2008, 67, 248–250. [Google Scholar] [CrossRef] [PubMed]
- Park, J.H.; Kim, Y.J.; Park, B.L.; Bae, J.S.; Shin, H.D.; Bae, S.-C. Lack of Association between Interleukin 23 Receptor Gene Polymorphisms and Rheumatoid Arthritis Susceptibility. Rheumatol. Int. 2009, 29, 781–786. [Google Scholar] [CrossRef] [PubMed]
- Hamdy, G.; Darweesh, H.; Khattab, E.A.; Fawzy, S.; Fawzy, E.; Sheta, M. Evidence of Association of Interleukin-23 Receptor Gene Polymorphisms with Egyptian Rheumatoid Arthritis Patients. Hum. Immunol. 2015, 76, 417–420. [Google Scholar] [CrossRef] [PubMed]
- Tagiev, A.F.; Surin, V.L.; Osokina, A.V.; Luk’ianenko, A.V.; Smirnova, O.V.; Tsetaeva, N.V.; Mikhaĭlova, E.A.; Isaev, V.G.; Grineva, N.I. Polymorphism at codon 117 of the granulocyte-macrophage colony-stimulating factor (GM-CSF) gene. Genetika 1995, 31, 1370–1374. [Google Scholar] [PubMed]
- He, J.-Q.; Jian, R.; Moira, C.-Y.; Allan, B.; Helen, D.-W.; Peter, P.; Andrew, S. Polymorphisms of the GM-CSF Genes and the Development of Atopic Diseases in at-Risk Children. Chest 2003, 123, 438S. [Google Scholar] [CrossRef] [PubMed]
- Rafatpanah, H.; Bennett, E.; Pravica, V.; McCoy, M.J.; David, T.J.; Hutchinson, I.V.; Arkwright, P.D. Association between Novel GM-CSF Gene Polymorphisms and the Frequency and Severity of Atopic Dermatitis. J. Allergy Clin. Immunol. 2003, 112, 593–598. [Google Scholar] [CrossRef]
- Saeki, H.; Tsunemi, Y.; Asano, N.; Nakamura, K.; Sekiya, T.; Hirai, K.; Kakinuma, T.; Fujita, H.; Kagami, S.; Tamaki, K. Analysis of GM-CSF Gene Polymorphisms (3606T/C and 3928C/T) in Japanese Patients with Atopic Dermatitis. Clin. Exp. Dermatol. 2006, 31, 278–280. [Google Scholar] [CrossRef]
- Wilkowska, A.; Gleń, J.; Zabłotna, M.; Trzeciak, M.; Ryduchowska, M.; Sobjanek, M.; Nedoszytko, B.; Nowicki, R.; Sokołowska-Wojdyło, M. The Association of GM-CSF-677A/C Promoter Gene Polymorphism with the Occurrence and Severity of Atopic Dermatitis in a Polish Population. Int. J. Dermatol. 2014, 53, e172–e174. [Google Scholar] [CrossRef]
- Abdelaal, E.B.; Abdelsamie, H.M.; Attia, S.M.; Amr, K.S.; Eldahshan, R.M.; Elsaie, M.L. Association of a Novel Granulocyte-Macrophage Colony-Stimulating Factor (GM-CSF)-3928C/T and GM-CSF(3606T⁄C) Promoter Gene Polymorphisms with the Pathogenesis and Severity of Acne Vulgaris: A Case-Controlled Study. J. Cosmet. Dermatol. 2021, 20, 3679–3683. [Google Scholar] [CrossRef]
- Assmann, G.; Koenig, J.; Pfreundschuh, M.; Epplen, J.T.; Kekow, J.; Roemer, K.; Wieczorek, S. Genetic Variations in Genes Encoding RANK, RANKL, and OPG in Rheumatoid Arthritis: A Case-Control Study. J. Rheumatol. 2010, 37, 900–904. [Google Scholar] [CrossRef]
- Xu, S.; Ma, X.-X.; Hu, L.-W.; Peng, L.-P.; Pan, F.-M.; Xu, J.-H. Single Nucleotide Polymorphism of RANKL and OPG Genes May Play a Role in Bone and Joint Injury in Rheumatoid Arthritis. Clin. Exp. Rheumatol. 2014, 32, 697–704. [Google Scholar]
- Yang, H.; Liu, W.; Zhou, X.; Rui, H.; Zhang, H.; Liu, R. The Association between RANK, RANKL and OPG Gene Polymorphisms and the Risk of Rheumatoid Arthritis: A Case-Controlled Study and Meta-Analysis. Biosci. Rep. 2019, 39, BSR20182356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdi, S.; Bukhari, I.; Ansari, M.G.A.; BinBaz, R.A.; Mohammed, A.K.; Hussain, S.D.; Aljohani, N.; Al-Daghri, N.M. Association of Polymorphisms in RANK and RANKL Genes with Osteopenia in Arab Postmenopausal Women. Dis. Markers 2020, 2020, 1285216. [Google Scholar] [CrossRef] [PubMed]
- Wielińska, J.; Kolossa, K.; Świerkot, J.; Dratwa, M.; Iwaszko, M.; Bugaj, B.; Wysoczańska, B.; Chaszczewska-Markowska, M.; Jeka, S.; Bogunia-Kubik, K. Polymorphisms within the RANK and RANKL Encoding Genes in Patients with Rheumatoid Arthritis: Association with Disease Progression and Effectiveness of the Biological Treatment. Arch. Immunol. Ther. Exp. 2020, 68, 24. [Google Scholar] [CrossRef] [PubMed]
- Łacina, P.; Butrym, A.; Humiński, M.; Dratwa, M.; Frontkiewicz, D.; Mazur, G.; Bogunia-Kubik, K. Association of RANK and RANKL Gene Polymorphism with Survival and Calcium Levels in Multiple Myeloma. Mol. Carcinog. 2021, 60, 106–112. [Google Scholar] [CrossRef] [PubMed]
- Abdi, S.; Binbaz, R.A.; Mohammed, A.K.; Ansari, M.G.A.; Wani, K.; Amer, O.E.; Alnaami, A.M.; Aljohani, N.; Al-Daghri, N.M. Association of RANKL and OPG Gene Polymorphism in Arab Women with and without Osteoporosis. Genes 2021, 12, 200. [Google Scholar] [CrossRef] [PubMed]
- Gabay, C.; Lamacchia, C.; Palmer, G. IL-1 Pathways in Inflammation and Human Diseases. Nat. Rev. Rheumatol. 2010, 6, 232–241. [Google Scholar] [CrossRef]
- Schett, G.; Dayer, J.-M.; Manger, B. Interleukin-1 Function and Role in Rheumatic Disease. Nat. Rev. Rheumatol. 2016, 12, 14–24. [Google Scholar] [CrossRef] [PubMed]
- Alten, R.; Gomez-Reino, J.; Durez, P.; Beaulieu, A.; Sebba, A.; Krammer, G.; Preiss, R.; Arulmani, U.; Widmer, A.; Gitton, X.; et al. Efficacy and Safety of the Human Anti-IL-1beta Monoclonal Antibody Canakinumab in Rheumatoid Arthritis: Results of a 12-Week, Phase II, Dose-Finding Study. BMC Musculoskelet. Disord. 2011, 12, 153. [Google Scholar] [CrossRef] [PubMed]
- Geiler, J.; McDermott, M.F. Gevokizumab, an Anti-IL-1β MAb for the Potential Treatment of Type 1 and 2 Diabetes, Rheumatoid Arthritis and Cardiovascular Disease. Curr. Opin. Mol. Ther. 2010, 12, 755–769. [Google Scholar] [PubMed]
- Miossec, P.; Korn, T.; Kuchroo, V.K. Interleukin-17 and Type 17 Helper T Cells. N. Engl. J. Med. 2009, 361, 888–898. [Google Scholar] [CrossRef] [Green Version]
- Weaver, C.T.; Hatton, R.D.; Mangan, P.R.; Harrington, L.E. IL-17 Family Cytokines and the Expanding Diversity of Effector T Cell Lineages. Annu. Rev. Immunol. 2007, 25, 821–852. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaffen, S.L.; Jain, R.; Garg, A.V.; Cua, D.J. The IL-23-IL-17 Immune Axis: From Mechanisms to Therapeutic Testing. Nat. Rev. Immunol. 2014, 14, 585–600. [Google Scholar] [CrossRef] [PubMed]
- Noack, M.; Miossec, P. Selected Cytokine Pathways in Rheumatoid Arthritis. Semin. Immunopathol. 2017, 39, 365–383. [Google Scholar] [CrossRef] [PubMed]
- Kunwar, S.; Dahal, K.; Sharma, S. Anti-IL-17 Therapy in Treatment of Rheumatoid Arthritis: A Systematic Literature Review and Meta-Analysis of Randomized Controlled Trials. Rheumatol. Int. 2016, 36, 1065–1075. [Google Scholar] [CrossRef]
- Wu, D.; Hou, S.-Y.; Zhao, S.; Hou, L.-X.; Jiao, T.; Xu, N.-N.; Zhang, N. Meta-Analysis of IL-17 Inhibitors in Two Populations of Rheumatoid Arthritis Patients: Biologic-Naïve or Tumor Necrosis Factor Inhibitor Inadequate Responders. Clin. Rheumatol. 2019, 38, 2747–2756. [Google Scholar] [CrossRef]
- Huang, Y.; Fan, Y.; Liu, Y.; Xie, W.; Zhang, Z. Efficacy and Safety of Secukinumab in Active Rheumatoid Arthritis with an Inadequate Response to Tumor Necrosis Factor Inhibitors: A Meta-Analysis of Phase III Randomized Controlled Trials. Clin. Rheumatol. 2019, 38, 2765–2776. [Google Scholar] [CrossRef]
- Martin, D.A.; Churchill, M.; Flores-Suarez, L.; Cardiel, M.H.; Wallace, D.; Martin, R.; Phillips, K.; Kaine, J.L.; Dong, H.; Salinger, D.; et al. A Phase Ib Multiple Ascending Dose Study Evaluating Safety, Pharmacokinetics, and Early Clinical Response of Brodalumab, a Human Anti-IL-17R Antibody, in Methotrexate-Resistant Rheumatoid Arthritis. Arthritis Res. Ther. 2013, 15, R164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Glatt, S.; Taylor, P.C.; McInnes, I.B.; Schett, G.; Landewé, R.; Baeten, D.; Ionescu, L.; Strimenopoulou, F.; Watling, M.I.L.; Shaw, S. Efficacy and Safety of Bimekizumab as Add-on Therapy for Rheumatoid Arthritis in Patients with Inadequate Response to Certolizumab Pegol: A Proof-of-Concept Study. Ann. Rheum. Dis. 2019, 78, 1033–1040. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kleinschek, M.A.; Muller, U.; Brodie, S.J.; Stenzel, W.; Kohler, G.; Blumenschein, W.M.; Straubinger, R.K.; McClanahan, T.; Kastelein, R.A.; Alber, G. IL-23 Enhances the Inflammatory Cell Response in Cryptococcus Neoformans Infection and Induces a Cytokine Pattern Distinct from IL-12. J. Immunol. 2006, 176, 1098–1106. [Google Scholar] [CrossRef] [Green Version]
- Bunte, K.; Beikler, T. Th17 Cells and the IL-23/IL-17 Axis in the Pathogenesis of Periodontitis and Immune-Mediated Inflammatory Diseases. Int. J. Mol. Sci. 2019, 20, 3394. [Google Scholar] [CrossRef] [Green Version]
- Luo, J.; Wu, S.-J.; Lacy, E.R.; Orlovsky, Y.; Baker, A.; Teplyakov, A.; Obmolova, G.; Heavner, G.A.; Richter, H.-T.; Benson, J. Structural Basis for the Dual Recognition of IL-12 and IL-23 by Ustekinumab. J. Mol. Biol. 2010, 402, 797–812. [Google Scholar] [CrossRef] [PubMed]
- Machado, Á.; Torres, T. Guselkumab for the Treatment of Psoriasis. BioDrugs Clin. Immunother. Biopharm. Gene Ther. 2018, 32, 119–128. [Google Scholar] [CrossRef]
- Smolen, J.S.; Agarwal, S.K.; Ilivanova, E.; Xu, X.L.; Miao, Y.; Zhuang, Y.; Nnane, I.; Radziszewski, W.; Greenspan, A.; Beutler, A.; et al. A Randomised Phase II Study Evaluating the Efficacy and Safety of Subcutaneously Administered Ustekinumab and Guselkumab in Patients with Active Rheumatoid Arthritis despite Treatment with Methotrexate. Ann. Rheum. Dis. 2017, 76, 831–839. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kondo, N.; Kuroda, T.; Kobayashi, D. Cytokine Networks in the Pathogenesis of Rheumatoid Arthritis. Int. J. Mol. Sci. 2021, 22, 10922. [Google Scholar] [CrossRef]
- Haworth, C.; Brennan, F.M.; Chantry, D.; Turner, M.; Maini, R.N.; Feldmann, M. Expression of Granulocyte-Macrophage Colony-Stimulating Factor in Rheumatoid Arthritis: Regulation by Tumor Necrosis Factor-Alpha. Eur. J. Immunol. 1991, 21, 2575–2579. [Google Scholar] [CrossRef] [PubMed]
- Crotti, C.; Biggioggero, M.; Becciolini, A.; Agape, E.; Favalli, E.G. Mavrilimumab: A Unique Insight and Update on the Current Status in the Treatment of Rheumatoid Arthritis. Expert Opin. Investig. Drugs 2019, 28, 573–581. [Google Scholar] [CrossRef] [PubMed]
- Di Franco, M.; Gerardi, M.C.; Lucchino, B.; Conti, F. Mavrilimumab: An Evidence Based Review of Its Potential in the Treatment of Rheumatoid Arthritis. Core Evid. 2014, 9, 41–48. [Google Scholar] [CrossRef] [Green Version]
- Anderson, D.M.; Maraskovsky, E.; Billingsley, W.L.; Dougall, W.C.; Tometsko, M.E.; Roux, E.R.; Teepe, M.C.; DuBose, R.F.; Cosman, D.; Galibert, L. A Homologue of the TNF Receptor and Its Ligand Enhance T-Cell Growth and Dendritic-Cell Function. Nature 1997, 390, 175–179. [Google Scholar] [CrossRef]
- Tanaka, S.; Tanaka, Y. RANKL as a Therapeutic Target of Rheumatoid Arthritis. J. Bone Miner. Metab. 2021, 39, 106–112. [Google Scholar] [CrossRef]
- Chiu, Y.G.; Ritchlin, C.T. Denosumab: Targeting the RANKL Pathway to Treat Rheumatoid Arthritis. Expert Opin. Biol. Ther. 2017, 17, 119–128. [Google Scholar] [CrossRef] [Green Version]
- Maranini, B.; Bortoluzzi, A.; Silvagni, E.; Govoni, M. Focus on Sex and Gender: What We Need to Know in the Management of Rheumatoid Arthritis. J. Pers. Med. 2022, 12, 499. [Google Scholar] [CrossRef] [PubMed]
- Bellando-Randone, S.; Russo, E.; Venerito, V.; Matucci-Cerinic, M.; Iannone, F.; Tangaro, S.; Amedei, A. Exploring the Oral Microbiome in Rheumatic Diseases, State of Art and Future Prospective in Personalized Medicine with an AI Approach. J. Pers. Med. 2021, 11, 625. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, Y.; Kanayama, N.; Nakayama, Y.; Matsushima, N. Current Status, Issues and Future Prospects of Personalized Medicine for Each Disease. J. Pers. Med. 2022, 12, 444. [Google Scholar] [CrossRef] [PubMed]
| Biological Agent | Gene | Polymorphism | Clinical Outcome(s) | Refs. |
|---|---|---|---|---|
| Adalimumab | FCGR2A | rs1801274 | Significantly associated with the clinical response | [39] |
| DHX32 | rs12356233 | |||
| RGS12 | rs4690093 | Nominally significantly associated with the response | ||
| IL-6 | rs1800795 (−174 G/C) | Significantly associated with a better response | [40] | |
| PTPN22 | 1858 C/T | No effect on efficacy adalimumab | [41] | |
| TNF | rs1800629 (−308 G/A) | TNF-308 G/G genotype associated with better clinical effect than TNF −308 A/G | [42,43,44,45] | |
| −238A/G, −308A/G and −857C/T | TNF-α locus haplotype (−238G/−308G/−857C) was associated with a lower response | [46] | ||
| TNFR2 | 676 T/G | TNFR2 676 T/T genotype associated with a better clinical response than TNFR2 676 T/G | [47] |
| Biological Agent | Gene | Polymorphism | Clinical Outcome(s) | Refs. |
|---|---|---|---|---|
| Infliximab | FCGR2A | rs1801274 | No effect on response to infliximab | [39] |
| H131R | FCGR2A-RR was associated with a better response compared to RH or HH | [78] | ||
| FCGR3A | V158F | FCGR3A-FF was associated with a better response compared to VV or VF | ||
| RGS12 | rs2857859 | Nominally significantly associated with the response | [39] | |
| PTPN22 | 1858 C/T | No effect on response to infliximab | [41] | |
| MHC | MHC polymorphisms | TNFa11;b4 associated with responders | [79] | |
| TNF | rs1800629 (−308 G/A) | TNF-308 G/G associated with a better response than TNF −308 A/A or A/G | [61,63,64] | |
| −308 G/A | No effect on response to infliximab | [67] | ||
| −308 G/A, −238 G/A, 489 G/A, −857 C/T | [70] | |||
| rs361525 (−238 G/A) | TNF −238G/A was associated with poorer response | [71] | ||
| −238 G/G, +489 A/A | TNF −238 G/G was associated with severe RA | [80] | ||
| −308 G/A, −238 G/A | No effect on response to infliximab | [81] | ||
| TNFR1 | 36 A/A, 676 T/G | TNFR1A 36 A/A was associated with better response compared to G/G and TNFR1B 676 T/G was not associated with response to infliximab | ||
| 36 A/G | No effect on response to infliximab | [70] | ||
| TNFR2 | 676 T/G | A combination of 676 T/G (TNFR2) and −857 C/T (TNF-α) could be used for prognosis of clinical response to infliximab |
| Biological Agent | Gene | Polymorphism | Clinical Outcome(s) | Refs. |
|---|---|---|---|---|
| Rituximab | FCGR2A | rs1801274 | FCGR2A rs1801274-T/T genotype was associated with better clinical response to rituximab | [96] |
| FCGR3A | rs396991 | FCGR3A rs396991-G allele genotype was associated with better response to rituximab | ||
| −158 V/F | FCGR3A −158 V/V was associated with a better response than V/F or F/F | [97] | ||
| FCGR3A −158 V allele was independently associated with better response to rituximab | [98] | |||
| FCGR3A −158 V/V and V/F was associated with a better response than F/F | [99] | |||
| FCGR3A −158 V/F was not associated with CDAI response to rituximab | [100] | |||
| BAFF | −871 C/T | BAFF −871 C/C was associated with a better response than T/T | [101] | |
| IL-6 | rs1800795 (−174 G/C) | IL-6 −174 GG/GC genotypes was associated with better responses than with −174 C/C genotypes | [102] | |
| TGFβ1 | rs1800470, rs1800471 | TGFβ1 SNPs was associated with good response to rituximab | [103] |
| Biological Agent | Gene | Polymorphism | Clinical Outcome(s) | Refs. |
|---|---|---|---|---|
| Tocilizumab | FCGR2A | rs1801274 | Not associated with response to tocilizumab | [96] |
| FCGR3A | rs396991 | FCGR3A rs396991-T/T genotype is associated with better EULAR response to tocilizumab | ||
| HLA-DRB1 | rs11052877, rs4910008, rs9594987, rs10108210, rs703297, rs703505, rs1560011, rs7055107 | The shared epitope HLA-DRB1 had no association with tocilizumab response | [110] | |
| IL-6R | rs12083537, rs2228145, rs4329505, rs11265618 | rs12083537-A/A and rs11265618-C/C were associated with higher LDA rates | [111] |
| Biological Agent | Polymorphism | Clinical Outcome(s) | Refs. |
|---|---|---|---|
| Canakinumab, Gevokizumab | IL-1α | rs17561 | [115] |
| IL-1β | rs16944 | [115] | |
| rs16944, rs1143634 | [116] | ||
| [117] | |||
| [118] | |||
| [119] | |||
| IL-1Ra (IL-1RN) | 2018 C/T | [117] | |
| 5111 T/C, 2017 T/C | [115] | ||
| Secukinumab, Ixekizumab, Bimekizumab | IL-17A | rs4711998, rs8193036, rs3819024, rs2275913, rs7747909 | [120] |
| rs2275913, rs3804513, rs3748067, rs1974226 | [121] | ||
| rs2275913 | [122] | ||
| [123] | |||
| rs2275913, rs3819024, rs3819025, rs4711998, rs8193036, rs8193037, rs3804513 | [124] | ||
| rs2275913, rs3804513 | [125] | ||
| rs2275913 | [126] | ||
| [127] | |||
| [128] | |||
| [129] | |||
| [130] | |||
| [131] | |||
| rs2275913, rs3819024, rs4711998, rs8193036 | [132] | ||
| rs2275913 | [133] | ||
| [134] | |||
| [135] | |||
| [136] | |||
| [137] | |||
| [138] | |||
| IL-17F | rs763780, rs2397084 | [121] | |
| rs763780 | [122] | ||
| rs763780, rs2397084 | [123] | ||
| rs763780 | [125] | ||
| [126] | |||
| rs763780, rs11465553, rs2397084 | [127] | ||
| rs763780, rs2397084 | [128] | ||
| [129] | |||
| rs763780 | [130] | ||
| rs763780, rs2397084 | [132] | ||
| [134] | |||
| rs763780 | [135] | ||
| rs763780, rs2397084 | [136] | ||
| rs763780 | [138] | ||
| Brodalumab | IL-17RC | rs708567 | [133] |
| Ustekinumab, Guselkumab | IL-23R | rs11209026, rs134315, rs10489629, rs7517847 | [125] |
| rs11209026, rs1343151, rs10489629 | [128] | ||
| rs10889677 | [130] | ||
| rs1004819, rs10489629, rs11209026, rs1343151, rs10889677, rs11209032, rs1495965 | [139] | ||
| rs10889677, rs2201841, rs1884444 | [140] | ||
| rs1004819, rs7517847, rs10489629, rs2201841, rs1343151, rs11209032, rs1495965 | [141] | ||
| rs11209026, rs2201841, rs10889677 | [142] | ||
| Lenzilumab, Namilumab, Otilimab, Gimsilumab | GM-CSF | 177 T/C | [143] |
| 545 G/A, 3606 T/C, 3928 C/T | [144] | ||
| 677 C/C | [145] | ||
| 3606 T/C, 3928 C/T | [146] | ||
| 677 A/C | [147] | ||
| 3928 C/T, 3606 T/C | [148] | ||
| Denosumab | RANKL | rs9533156, rs2277438, rs1054016 | [149] |
| rs2277438 | [150] | ||
| rs2277438, rs7984870 | [151] | ||
| rs2277438, rs9533156 | [152] | ||
| rs7325635, rs7988338 | [153] | ||
| [154] | |||
| rs2277438, rs9533156 | [155] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lim, S.H.; Kim, K.; Choi, C.-I. Pharmacogenomics of Monoclonal Antibodies for the Treatment of Rheumatoid Arthritis. J. Pers. Med. 2022, 12, 1265. https://doi.org/10.3390/jpm12081265
Lim SH, Kim K, Choi C-I. Pharmacogenomics of Monoclonal Antibodies for the Treatment of Rheumatoid Arthritis. Journal of Personalized Medicine. 2022; 12(8):1265. https://doi.org/10.3390/jpm12081265
Chicago/Turabian StyleLim, Sung Ho, Khangyoo Kim, and Chang-Ik Choi. 2022. "Pharmacogenomics of Monoclonal Antibodies for the Treatment of Rheumatoid Arthritis" Journal of Personalized Medicine 12, no. 8: 1265. https://doi.org/10.3390/jpm12081265