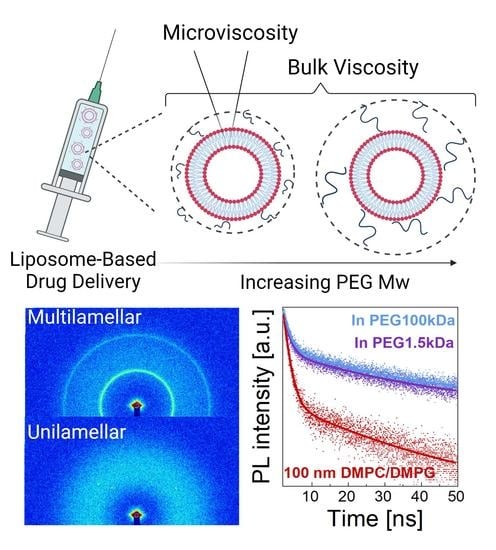
Multiscale Dynamics of Lipid Vesicles in Polymeric Microenvironment
Abstract
:1. Introduction
2. Materials and Methods
2.1. Preparation of Liposome Solutions
2.2. Preparation of Liposome Solutions Containing Oil Red O
2.3. Preparation of Liposome–Polymer Mixtures
2.4. Dynamic Light Scattering (DLS)
2.5. Differential Scanning Calorimetry (DSC)
2.6. Rheology
2.7. Small-Angle X-ray Scattering (SAXS)
2.8. Cryo-TEM
2.9. UV-Vis Spectroscopy
2.10. Time-Resolved Photoluminescence
3. Results and Discussion
3.1. Structure
3.2. Dynamics
3.2.1. Bulk Diffusion of Vesicles
3.2.2. Macroscopic Viscosity
3.2.3. Microscopic Viscosity in Bilayers
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Akbarzadeh, A.; Rezaei-Sadabady, R.; Davaran, S.; Joo, S.W.; Zarghami, N.; Hanifehpour, Y.; Samiei, M.; Kouhi, M.; Nejati-Koshki, K. Liposome: Classification, preparation, and applications. Nanoscale Res. Lett. 2013, 8, 102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, M.; Du, C.; Guo, N.; Teng, Y.; Meng, X.; Sun, H.; Li, S.; Yu, P.; Galons, H. Composition design and medical application of liposomes. Eur. J. Med. Chem. 2019, 164, 640–653. [Google Scholar] [CrossRef] [PubMed]
- Yeagle, P.L. Non-covalent binding of membrane lipids to membrane proteins. Biochim. et Biophys. Acta (BBA) Biomembr. 2014, 1838, 1548–1559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, S.; Gao, H.; Bao, G. Physical Principles of Nanoparticle Cellular Endocytosis. ACS Nano 2015, 9, 8655–8671. [Google Scholar] [CrossRef] [Green Version]
- Fujiwara, T.; Ritchie, K.; Murakoshi, H.; Jacobson, K.; Kusumi, A. Phospholipids undergo hop diffusion in compartmentalized cell membrane. J. Cell Biol. 2002, 157, 1071–1082. [Google Scholar] [CrossRef] [Green Version]
- Turecek, P.L.; Siekmann, J. 4-PEG-protein conjugates: Nonclinical and clinical toxicity considerations. In Polymer-Protein Conjugates; Pasut, G., Zalipsky, S., Eds.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 61–101. [Google Scholar]
- Jesorka, A.; Orwar, O. Liposomes: Technologies and Analytical Applications. Annu. Rev. Anal. Chem. 2008, 1, 801–832. [Google Scholar] [CrossRef]
- Kazakov, S. Liposome-Nanogel Structures for Future Pharmaceutical Applications: An Updated Review. Curr. Pharm. Des. 2016, 22, 1391–1413. [Google Scholar] [CrossRef]
- Birgul Akolpoglu, M.; Inceoglu, Y.; Kizilel, S. An all-aqueous approach for physical immobilization of PEG-lipid microgels on organoid surfaces. Colloids Surf. B Biointerfaces 2020, 186, 110708. [Google Scholar] [CrossRef]
- Nanomedicine and the COVID-19 vaccines. Nat. Nanotechnol. 2020, 15, 963. [CrossRef]
- Bigini, P.; Gobbi, M.; Bonati, M.; Clavenna, A.; Zucchetti, M.; Garattini, S.; Pasut, G. The role and impact of polyethylene glycol on anaphylactic reactions to COVID-19 nano-vaccines. Nat. Nanotechnol. 2021, 16, 1169–1171. [Google Scholar] [CrossRef]
- Sellaturay, P.; Nasser, S.; Islam, S.; Gurugama, P.; Ewan, P.W. Polyethylene glycol (PEG) is a cause of anaphylaxis to the Pfizer/BioNTech mRNA COVID-19 vaccine. Clin. Exp. Allergy 2021, 51, 861–863. [Google Scholar] [CrossRef]
- Granek, R.; Diamant, H. Membrane undulations in a structured fluid: Universal dynamics at intermediate length and time scales. Eur. Phys. J. E 2018, 41, 1. [Google Scholar] [CrossRef] [Green Version]
- Elmore, D.E. Molecular dynamics simulation of a phosphatidylglycerol membrane. FEBS Lett. 2005, 580, 144–148. [Google Scholar] [CrossRef] [Green Version]
- Dickey, A.; Faller, R. Examining the Contributions of Lipid Shape and Headgroup Charge on Bilayer Behavior. Biophys. J. 2008, 95, 2636–2646. [Google Scholar] [CrossRef] [Green Version]
- Bondar, A.-N.; White, S.H. Hydrogen bond dynamics in membrane protein function. Biochim. et Biophys. Acta Biomembr. 2012, 1818, 942–950. [Google Scholar] [CrossRef] [Green Version]
- Kozlowska, M.; Goclon, J.; Rodziewicz, P. Intramolecular Hydrogen Bonds in Low-Molecular-Weight Polyethylene Glycol. ChemPhysChem 2016, 17, 1143–1153. [Google Scholar] [CrossRef]
- Zhang, W.; Metzger, J.M.; Hackel, B.J.; Bates, F.S.; Lodge, T.P. Influence of the Headgroup on the Interaction of Poly(ethylene oxide)-Poly(propylene oxide) Block Copolymers with Lipid Bilayers. J. Phys. Chem. B 2020, 124, 2417–2424. [Google Scholar] [CrossRef]
- Pal, S.; Milano, G.; Roccatano, D. Synthetic Polymers and Biomembranes. How Do They Interact?: Atomistic Molecular Dynamics Simulation Study of PEO in Contact with a DMPC Lipid Bilayer. J. Phys. Chem. B 2006, 110, 26170–26179. [Google Scholar] [CrossRef]
- Lentz, B.R.; Lee, J.; Lentz, J.L.B.R. Poly(ethylene glycol) (PEG)-mediated fusion between pure lipid bilayers: A mechanism in common with viral fusion and secretory vesicle release? (Review). Mol. Membr. Biol. 1999, 16, 279–296. [Google Scholar] [CrossRef]
- Lentz, B.R. PEG as a tool to gain insight into membrane fusion. Eur. Biophys. J. 2006, 36, 315–326. [Google Scholar] [CrossRef]
- Dutheil, D.; Underhaug Gjerde, A.; Petit-Paris, I.; Mauco, G.; Holmsen, H. Polyethylene glycols interact with membrane glycerophospholipids: Is this part of their mechanism for hypothermic graft protection? J. Chem. Biol. 2009, 2, 39–49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalyanram, P.; Puri, A.; Gupta, A. Thermotropic effects of PEGylated lipids on the stability of HPPH-encapsulated lipid nanoparticles (LNP). J. Therm. Anal. 2021, 147, 6337–6348. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yang, L.-J.; Zhu, T.-T.; Wang, S.-Z.; Chen, Z.-X. Phase Transition of Phospholipid Vesicles Induced by Fatty Acids in Macromolecular Crowding: A Differential Scanning Calorimetry Study. Acta Phys.-Chim. Sin. 2016, 32, 2027–2038. [Google Scholar] [CrossRef]
- Yamazaki, M.; Ohshika, M.; Kashiwagi, N.; Asano, T. Phase transitions of phospholipid vesicles under osmotic stress and in the presence of ethylene glycol. Biophys. Chem. 1992, 43, 29–37. [Google Scholar] [CrossRef]
- Sahu, S.; Talele, P.; Patra, B.; Verma, R.S.; Mishra, A.K. A Multiparametric Fluorescence Probe to Understand the Physicochemical Properties of Small Unilamellar Lipid Vesicles in Poly(ethylene glycol)-Water Medium. Langmuir 2020, 36, 4842–4852. [Google Scholar] [CrossRef] [PubMed]
- Zakim, D.; Kavecansky, J.; Scarlata, S. Are membrane enzymes regulated by the viscosity of the membrane environ-ment? Biochemistry 1992, 31, 11589–11594. [Google Scholar] [CrossRef] [PubMed]
- Heron, D.S.; Shinitzky, M.; Hershkowitz, M.; Samuel, D. Lipid fluidity markedly modulates the binding of serotonin to mouse brain membranes. Proc. Natl. Acad. Sci. USA 1980, 77, 7463–7467. [Google Scholar] [CrossRef] [Green Version]
- Savigny, P.; Evans, J.; McGrath, K.M. Cell Membrane Structures during Exocytosis. Endocrinology 2007, 148, 3863–3874. [Google Scholar] [CrossRef] [Green Version]
- Grecco, H.E.; Schmick, M.; Bastiaens, P.I. Signaling from the Living Plasma Membrane. Cell 2011, 144, 897–909. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Escribá, P.V.; González-Ros, J.M.; Goñi, F.M.; Kinnunen, P.K.J.; Vigh, L.; Sánchez-Magraner, L.; Fernández, A.M.; Busquets, X.; Horváth, I.; Barceló-Coblijn, G. Membranes: A meeting point for lipids, proteins and therapies. J. Cell. Mol. Med. 2008, 12, 829–875. [Google Scholar] [CrossRef] [Green Version]
- Jing, H.; Wang, Y.; Desai, P.R.; Ramamurthi, K.S.; Das, S. Lipid flip-flop and desorption from supported lipid bilayers is independent of curvature. PLoS ONE 2020, 15, e0244460. [Google Scholar] [CrossRef] [PubMed]
- Marguet, D.; Lenne, P.-F.; Rigneault, H.; Hervé, R. Dynamics in the plasma membrane: How to combine fluidity and order. EMBO J. 2006, 25, 3446–3457. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Movileanu, L.; Popescu, D.; Ion, S.; Popescu, A.I. Transbilayer pores induced by thickness fluctuations. Bull. Math. Biol. 2006, 68, 1231–1255. [Google Scholar] [CrossRef] [PubMed]
- Eggert, U.S.; Mitchison, T.J.; Field, C.M. Animal Cytokinesis: From Parts List to Mechanisms. Annu. Rev. Biochem. 2006, 75, 543–566. [Google Scholar] [CrossRef] [PubMed]
- van Niel, G.; D’Angelo, G.; Raposo, G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213–228. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Benda, A.; Kwiatek, J.; Owen, D.M.; Gaus, K. Time-Resolved Laurdan Fluorescence Reveals Insights into Membrane Viscosity and Hydration Levels. Biophys. J. 2018, 115, 1498–1508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shinitzky, M.; Barenholz, Y. Dynamics of the hydrocarbon layer in liposomes of lecithin and sphingomyelin containing dicetylphosphate. J. Biol. Chem. 1974, 249, 2652–2657. [Google Scholar] [CrossRef]
- Kung, C.E.; Reed, J.K. Microviscosity measurements of phospholipid bilayers using fluorescent dyes that undergo torsional relaxation. Biochemistry 1986, 25, 6114–6121. [Google Scholar] [CrossRef]
- Nojima, Y.; Iwata, K. Viscosity Heterogeneity inside Lipid Bilayers of Single-Component Phosphatidylcholine Liposomes Observed with Picosecond Time-Resolved Fluorescence Spectroscopy. J. Phys. Chem. B 2014, 118, 8631–8641. [Google Scholar] [CrossRef]
- Bozuyuk, U.; Dogan, N.O.; Kizilel, S. Deep Insight into PEGylation of Bioadhesive Chitosan Nanoparticles: Sensitivity Study for the Key Parameters Through Artificial Neural Network Model. ACS Appl. Mater. Interfaces 2018, 10, 33945–33955. [Google Scholar] [CrossRef]
- Bozuyuk, U.; Gokulu, I.S.; Dogan, N.O.; Kizilel, S. A novel method for PEGylation of chitosan nanoparticles through photopolymerization. RSC Adv. 2019, 9, 14011–14015. [Google Scholar] [CrossRef] [Green Version]
- Macdonald, P.M.; Leisen, J.; Marassi, F.M. Response of phosphatidylcholine in the gel and liquid-crystalline states to membrane surface charges. Biochemistry 1991, 30, 3558–3566. [Google Scholar] [CrossRef]
- Kinnunen, P.K. On the principles of functional ordering in biological membranes. Chem. Phys. Lipids 1991, 57, 375–399. [Google Scholar] [CrossRef]
- Drazenovic, J.; Wang, H.; Roth, K.; Zhang, J.; Ahmed, S.; Chen, Y.; Bothun, G.; Wunder, S.L. Effect of lamellarity and size on calorimetric phase transitions in single component phosphatidylcholine vesicles. Biochim. et Biophys. Acta Biomembr. 2015, 1848, 532–543. [Google Scholar] [CrossRef] [Green Version]
- Heimburg, T. Mechanical aspects of membrane thermodynamics. Estimation of the mechanical properties of lipid membranes close to the chain melting transition from calorimetry. Biochim. Biophys. Acta Biomembr. 1998, 1415, 147–162. [Google Scholar] [CrossRef] [Green Version]
- Lichtenberg, D.; Menashe, M.; Donaldson, S.; Biltonen, R.L. Thermodynamic characterization of the pretrasition of unilamellar dipalmitoyl-phosphatidylcholine vesicles. Lipids 1984, 19, 395–400. [Google Scholar] [CrossRef]
- Konarev, P.V.; Gruzinov, A.Y.; Mertens, H.D.T.; Svergun, D.I. Restoring structural parameters of lipid mixtures from small-angle X-ray scattering data. J. Appl. Crystallogr. 2021, 54, 169–179. [Google Scholar] [CrossRef]
- Takeuchi, H.; Kojima H Fau-Yamamoto, H.; Yamamoto, H.; Fau-Kawashima, Y.; Kawashima, Y. Evaluation of circulation profiles of liposomes coated with hydrophilic polymers having different molecular weights in rats. J. Control. Release 2001, 75, 83–91. [Google Scholar] [CrossRef]
- Hammouda, B.; Ho, D.L.; Kline, S. Insight into Clustering in Poly(ethylene oxide) Solutions. Macromolecules 2004, 37, 6932–6937. [Google Scholar] [CrossRef]
- Doux, J.P.F.; Hall, B.A.; Killian, J.A. How lipid headgroups sense the membrane environment: An application of B№вЃґN NMR. Biophys. J. 2012, 103, 1245–1253. [Google Scholar] [CrossRef] [Green Version]
- De Mel, J.U.; Gupta, S.; Willner, L.; Allgaier, J.; Stingaciu, L.R.; Bleuel, M.; Schneider, G.J. Manipulating Phospholipid Vesicles at the Nanoscale: A Transformation from Unilamellar to Multilamellar by an n-Alkyl-poly(ethylene oxide). Langmuir 2021, 37, 2362–2375. [Google Scholar] [CrossRef] [PubMed]
- Devanand, K.; Selser, J.C. Asymptotic behavior and long-range interactions in aqueous solutions of poly(ethylene oxide). Macromolecules 1991, 24, 5943–5947. [Google Scholar] [CrossRef]
- Jouault, N.; Moll, J.F.; Meng, D.; Windsor, K.; Ramcharan, S.; Kearney, C.; Kumar, S.K. Bound Polymer Layer in Nanocomposites. ACS Macro Lett. 2013, 2, 371–374. [Google Scholar] [CrossRef] [PubMed]
- Bandara, S.R.; Molley, T.G.; Kim, H.; Bharath, P.A.; Kilian, K.A.; Leal, C. The structural fate of lipid nanoparticles in the extracellular matrix. Mater. Horizons 2019, 7, 125–134. [Google Scholar] [CrossRef]
- Money, N.P. Osmotic Pressure of Aqueous Polyethylene Glycols 1: Relationship between Molecular Weight and Vapor Pressure Deficit. Plant Physiol. 1989, 91, 766–769. [Google Scholar] [CrossRef] [PubMed]
- Falke, S.; Betzel, C. Dynamic Light Scattering (DLS): Principles, Perspectives, Applications to Biological Samples. Radiat. Bioanal. 2019, 8, 173–193. [Google Scholar]
- Enoki, T.A.; Henriques, V.B.; Lamy, M.T. Light scattering on the structural characterization of DMPG vesicles along the bilayer anomalous phase transition. Chem. Phys. Lipids 2012, 165, 826–837. [Google Scholar] [CrossRef]
- Gupta, S.; De Mel, J.U.; Schneider, G.J. Dynamics of liposomes in the fluid phase. Curr. Opin. Colloid Interface Sci. 2019, 42, 121–136. [Google Scholar] [CrossRef]
- Shinitzky, M. Membrane fluidity in malignancy Adversative and recuperative. Biochim. Biophys. Acta (BBA)-Rev. Cancer 1984, 738, 251–261. [Google Scholar] [CrossRef]
- Nakazawa, K.; Kawata, Y.; Hishida, M.; Yamamura, Y.; Saito, K. Reduction of Shear Viscosity in Phospholipid Vesicle Dispersions by Self-organized Ripple Structures of Vesicle Surfaces. Chem. Lett. 2018, 47, 240–242. [Google Scholar] [CrossRef]
- Fujii, S. Rheological characterization of thermal phase behavior of anionic lipid DMPG dispersions. J. Biorheol. 2017, 31, 6–11. [Google Scholar] [CrossRef] [Green Version]
- Krieger, I.M.; Dougherty, T.J. A Mechanism for Non-Newtonian Flow in Suspensions of Rigid Spheres. Trans. Soc. Rheol. 1959, 3, 137–152. [Google Scholar] [CrossRef]
- Transport Phenomena; Bird, R.B.; Stewart, W.E.; Lightfoot, E.N. (Eds.) John Wiley and Sons, Inc.: New York, NY, USA, 1960. [Google Scholar] [CrossRef]
- Shewan, H.M.; Stokes, J.R. Viscosity of soft spherical micro-hydrogel suspensions. J. Colloid Interface Sci. 2015, 442, 75–81. [Google Scholar] [CrossRef]
- Steinmark, I.E.; James, A.L.; Chung, P.-H.; Morton, P.E.; Parsons, M.; Dreiss, C.A.; Lorenz, C.D.; Yahioglu, G.; Suhling, K. Targeted fluorescence lifetime probes reveal responsive organelle viscosity and membrane fluidity. PLoS ONE 2019, 14, e0211165. [Google Scholar] [CrossRef]
- Raut, S.L.; Kimball, J.D.; Fudala, R.; Bora, I.; Chib, R.; Jaafari, H.; Castillo, M.K.; Smith, N.W.; Gryczynski, I.; Dzyuba, S.V.; et al. A triazine-based BODIPY trimer as a molecular viscometer. Phys. Chem. Chem. Phys. 2016, 18, 4535–4540. [Google Scholar] [CrossRef] [Green Version]
- Kuimova, M.K.; Yahioglu, G.; Levitt, J.A.; Suhling, K. Molecular Rotor Measures Viscosity of Live Cells via Fluorescence Lifetime Imaging. J. Am. Chem. Soc. 2008, 130, 6672–6673. [Google Scholar] [CrossRef] [Green Version]
- Mehlem, A.; Hagberg, C.; Muhl, L.; Eriksson, U.; Falkevall, A. Imaging of neutral lipids by oil red O for analyzing the metabolic status in health and disease. Nat. Protoc. 2013, 8, 1149–1154. [Google Scholar] [CrossRef] [Green Version]
- Förster, T.; Hoffmann, G. Die Viskositätsabhängigkeit der Fluoreszenzquantenausbeuten einiger Farbstoffsysteme. Z. Für Phys. Chem. 1971, 75, 63–76. [Google Scholar]
- Wu, Y.; Etefl, M.; Olzy Eska, A.; Hof, M.; Yahioglu, G.; Yip, P.; Casey, D.R.; Ces, O.; Humpolíčková, J.; Kuimova, M.K. Molecular rheometry: Direct determination of viscosity in Lo and Ld lipid phases via fluorescence lifetime imaging. Phys. Chem. Chem. Phys. 2013, 15, 14986–14993. [Google Scholar] [CrossRef] [Green Version]
- Someya, Y.; Yui, H. Fluorescence Lifetime Probe for Solvent Microviscosity Utilizing Anilinonaphthalene Sulfonate. Anal. Chem. 2010, 82, 5470–5476. [Google Scholar] [CrossRef]
- Macklin, J.J.; Trautman, J.K.; Harris, T.D.; Brus, L.E. Imaging and Time-Resolved Spectroscopy of Single Molecules at an Interface. Science 1996, 272, 255–258. [Google Scholar] [CrossRef]
- Mohapatra, M.; Mishra, A.K. Excited state proton transfer based fluorescent molecular probes and their application in studying lipid bilayer membranes. Photochem. Photobiol. Sci. 2019, 18, 2830–2848. [Google Scholar] [CrossRef] [PubMed]
- Merkel, R.; Sackmann, E.; Evans, E. Molecular friction and epitactic coupling between monolayers in supported bilayers. J. De Phys. 1989, 50, 1535–1555. [Google Scholar] [CrossRef]
- Nagao, M.; Kelley, E.G.; Faraone, A.; Saito, M.; Yoda, Y.; Kurokuzu, M.; Takata, S.; Seto, M.; Butler, P.D. Relationship between Viscosity and Acyl Tail Dynamics in Lipid Bilayers. Phys. Rev. Lett. 2021, 127, 078102. [Google Scholar] [CrossRef]
- Viguera, A.-R.; Alonso, A.; Goñi, F.M. Liposome aggregation induced by poly(ethylene glycol). Rapid kinetic studies. Colloids Surf. B Biointerfaces 1995, 3, 263–270. [Google Scholar] [CrossRef]
- Tilcock, C.P.; Fisher, D. The interaction of phospholipid membranes with poly(ethylene glycol). Vesicle aggregation and lipid exchange. Biochim. et Biophys. Acta Biomembr. 1982, 688, 645–652. [Google Scholar] [CrossRef]
- Fetters, L.J.; Lohse, D.J.; Richter, D.; Witten, T.A.; Zirkel, A. Connection between Polymer Molecular Weight, Density, Chain Dimensions, and Melt Viscoelastic Properties. Macromolecules 1994, 27, 4639–4647. [Google Scholar] [CrossRef]
- Kučerka, N.; Nieh, M.-P.; Katsaras, J. Fluid phase lipid areas and bilayer thicknesses of commonly used phosphatidylcholines as a function of temperature. Biochim. Biophys. Acta (BBA)-Biomembr. 2011, 1808, 2761–2771. [Google Scholar] [CrossRef]
- Drabik, D.; Chodaczek, G.; Kraszewski, S.; Langner, M. Mechanical Properties Determination of DMPC, DPPC, DSPC, and HSPC Solid-Ordered Bilayers. Langmuir 2020, 36, 3826–3835. [Google Scholar] [CrossRef]







| HD (nm) (35 °C) | PDI | Tc (°C) | Specific Enthalpy (J/g) | DT (m2/s) (35 °C) (x 10−12) | DT (m2/s) (15 °C) (x 10−12) | |
|---|---|---|---|---|---|---|
| Liposome only | 119.9 ± 1.11 | 0.075 ± 0.010 | 22.84 ± 0.11 | 0.335 ± 0.013 | 5.21 ± 0.018 | 3.73 ± 0.013 |
| In PEG 1.5 kDa | 121.4 ± 0.20 | 0.074 ± 0.009 | 23.55 ± 0.19 | 0.242 ± 0.015 | 5.12 ± 0.020 | 3.61 ± 0.015 |
| In PEG 20 kDa | 129.6 ± 1.10 | 0.077 ± 0.014 | 23.39 ± 0.02 | 0.241 ± 0.017 | 5.03 ± 0.023 | 3.58 ± 0.016 |
| In PEG 100 kDa | 135.7 ± 2.13 | 0.113 ± 0.022 | 23.95 ± 0.32 | 0.234 ± 0.020 | 5.02 ± 0.028 | 3.21 ± 0.020 |
| In PEG 400 kDa | 157.7 ± 3.28 | 0.150 ± 0.025 | 22.58 ± 0.07 | 0.256 ± 0.012 | 4.40 ± 0.019 | 2.72 ± 0.014 |
| τavg (ns) 35 °C | Microviscosity (cP) 35 °C | Bulk Viscosity (cP) 35 °C | τavg (ns) 15 °C | Microviscosity (cP) 15 °C | Bulk Viscosity (cP) 15 °C | |
|---|---|---|---|---|---|---|
| 100 nm liposomes | 24.01 ± 0.032 | 28.75 ± 0.85 | 1.414 | 28.91 ± 0.038 | 142.82 ± 13.54 | 1.546 |
| in PEG 1.5 kDa | 20.61 ± 0.013 | 1.02 ± 0.01 | 1.156 | 27.38 ± 0.033 | 3.36 ± 0.29 | 1.827 |
| in PEG 20 kDa | 21.43 ± 0.022 | 2.39 ± 0.05 | 3.406 | 27.59 ± 0.036 | 5.69 ± 0.53 | 4.163 |
| in PEG 100 kDa | 22.72 ± 0.019 | 8.59 ± 0.15 | 6.874 | 28.42 ± 0.029 | 43.93 ± 3.2 | 8.247 |
| in PEG 400 kDa | 23.91 ± 0.028 | 26.24 ± 0.68 | 23.94 | 28.74 ± 0.031 | 95.09 ± 7.34 | 53.35 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karaz, S.; Han, M.; Akay, G.; Onal, A.; Nizamoglu, S.; Kizilel, S.; Senses, E. Multiscale Dynamics of Lipid Vesicles in Polymeric Microenvironment. Membranes 2022, 12, 640. https://doi.org/10.3390/membranes12070640
Karaz S, Han M, Akay G, Onal A, Nizamoglu S, Kizilel S, Senses E. Multiscale Dynamics of Lipid Vesicles in Polymeric Microenvironment. Membranes. 2022; 12(7):640. https://doi.org/10.3390/membranes12070640
Chicago/Turabian StyleKaraz, Selcan, Mertcan Han, Gizem Akay, Asim Onal, Sedat Nizamoglu, Seda Kizilel, and Erkan Senses. 2022. "Multiscale Dynamics of Lipid Vesicles in Polymeric Microenvironment" Membranes 12, no. 7: 640. https://doi.org/10.3390/membranes12070640