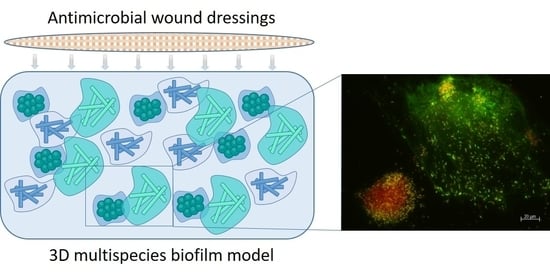
3D Biofilm Models Containing Multiple Species for Antimicrobial Testing of Wound Dressings
Abstract
:1. Introduction
2. Materials and Methods
2.1. Preparation of 3D Biofilm Models
2.2. Visualization of the 3D Biofilm
2.3. Stability Testing
2.4. Incubation with Wound Dressings
2.5. Quantification of the Antimicrobial Effect of Wound Dressings
2.6. Statistics
3. Results
3.1. Establishment of the 3D Biofilm Model
3.2. Visualization of the 3D Biofilm
3.3. Stability of the 3D Biofilm Model
3.4. Quantification of the Antimicrobial Effect of Wound Dressings
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bjarnsholt, T.; Mastroianni, E.; Kirketerp-Møller, K.; Stewart, P.S.; Mähr, A.M.; Domínguez Cabañes, A.; Nørager, R. The impact of mental models on the treatment and research of chronic infections due to biofilms. APMIS 2021, 129, 598–606. [Google Scholar] [CrossRef] [PubMed]
- Schultz, G.; Bjarnsholt, T.; James, G.A.; Leaper, D.J.; McBain, A.J.; Malone, M.; Stoodley, P.; Swanson, T.; Tachi, M.; Wolcott, R.D. Consensus guidelines for the identification and treatment of biofilms in chronic nonhealing wounds. Wound Repair Regen. 2017, 25, 744–757. [Google Scholar] [CrossRef] [PubMed]
- Demidova-Rice, T.N.; Hamblin, M.R.; Herman, I.M. Acute and impaired wound healing: Pathophysiology and current methods for drug delivery, part 1: Normal and chronic wounds: Biology, causes, and approaches to care. Adv. Ski. Wound Care 2012, 25, 304–314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haalboom, M. Chronic Wounds: Innovations in Diagnostics and Therapeutics. Curr. Med. Chem. 2018, 25, 5772–5781. [Google Scholar] [CrossRef]
- Heyer, K.; Herberger, K.; Protz, K.; Glaeske, G.; Augustin, M. Epidemiology of chronic wounds in Germany: Analysis of statutory health insurance data. Wound Repair Regen. 2016, 24, 434–442. [Google Scholar] [CrossRef]
- Percival, S.L.; McCarty, S.M.; Lipsky, B. Biofilms and Wounds: An Overview of the Evidence. Adv. Wound Care 2015, 4, 373–381. [Google Scholar] [CrossRef] [Green Version]
- Malone, M.; Bjarnsholt, T.; McBain, A.J.; James, G.A.; Stoodley, P.; Leaper, D.; Tachi, M.; Schultz, G.; Swanson, T.; Wolcott, R.D. The prevalence of biofilms in chronic wounds: A systematic review and meta-analysis of published data. J. Wound Care 2017, 26, 20–25. [Google Scholar] [CrossRef] [Green Version]
- Kirketerp-Møller, K.; Jensen, P.Ø.; Fazli, M.; Madsen, K.G.; Pedersen, J.; Moser, C.; Tolker-Nielsen, T.; Høiby, N.; Givskov, M.; Bjarnsholt, T. Distribution, Organization, and Ecology of Bacteria in Chronic Wounds. J. Clin. Microbiol. 2008, 46, 2717. [Google Scholar] [CrossRef] [Green Version]
- Johani, K.; Malone, M.; Jensen, S.; Gosbell, I.; Dickson, H.; Hu, H.; Vickery, K. Microscopy visualisation confirms multi-species biofilms are ubiquitous in diabetic foot ulcers. Int. Wound J. 2017, 14, 1160–1169. [Google Scholar] [CrossRef]
- James, G.A.; Swogger, E.; Wolcott, R.; Pulcini, E.; Secor, P.; Sestrich, J.; Costerton, J.W.; Stewart, P.S. Biofilms in chronic wounds. Wound Repair Regen. 2008, 16, 37–44. [Google Scholar] [CrossRef]
- Bjarnsholt, T.; Alhede, M.; Alhede, M.; Eickhardt-Sørensen, S.R.; Moser, C.; Kühl, M.; Jensen, P.; Høiby, N. The in vivo biofilm. Trends Microbiol. 2013, 21, 466–474. [Google Scholar] [CrossRef] [PubMed]
- Wiegand, C.; Tittelbach, J.; Hipler, C.; Elsner, P. Clinical efficacy of dressings for treatment of heavily exuding chronic wounds. Chronic Wound Care Manag. Res. 2015, 2, 11. [Google Scholar] [CrossRef] [Green Version]
- Gerber, V. Wundauflagen—was für wen? Allgemeinarzt. 2007, 11, 37–39. [Google Scholar]
- Rezvani Ghomi, E.; Khalili, S.; Nouri Khorasani, S.; Esmaeely Neisiany, R.; Ramakrishna, S. Wound dressings: Current advances and future directions. J. Appl. Polym. Sci. 2019, 136, 47738. [Google Scholar] [CrossRef] [Green Version]
- Farahani, M.; Shafiee, A. Wound Healing: From Passive to Smart Dressings. Adv. Healthc. Mater. 2021, 10, e2100477. [Google Scholar] [CrossRef]
- Thaarup, I.C.; Bjarnsholt, T. Current In Vitro Biofilm-Infected Chronic Wound Models for Developing New Treatment Possibilities. Adv Wound Care 2021, 10, 91–102. [Google Scholar] [CrossRef]
- Bahamondez-Canas, T.F.; Heersema, L.A.; Smyth, H.D.C. Current Status of In Vitro Models and Assays for Susceptibility Testing for Wound Biofilm Infections. Biomedicines 2019, 7, 34. [Google Scholar] [CrossRef] [Green Version]
- Brackman, G.; Coenye, T. In Vitro and In Vivo Biofilm Wound Models and Their Application. Adv. Exp. Med. Biol. 2016, 897, 15–32. [Google Scholar] [CrossRef]
- Coenye, T.; Nelis, H.J. In vitro and in vivo model systems to study microbial biofilm formation. J. Microbiol. Meth. 2010, 83, 89–105. [Google Scholar] [CrossRef]
- Percival, S.L.; Hill, K.E.; Williams, D.W.; Hooper, S.J.; Thomas, D.W.; Costerton, J.W. A review of the scientific evidence for biofilms in wounds. Wound Repair Regen. 2012, 20, 647–657. [Google Scholar] [CrossRef]
- Vyas, H.K.N.; Xia, B.; Mai-Prochnow, A. Clinically relevant in vitro biofilm models: A need to mimic and recapitulate the host environment. Biofilm 2022, 4, 100069. [Google Scholar] [CrossRef]
- Percival, S.L.; Mayer, D.; Salisbury, A.-M. Efficacy of a surfactant-based wound dressing on biofilm control. Wound Repair Regen. 2017, 25, 767–773. [Google Scholar] [CrossRef] [PubMed]
- Gawande, P.V.; Clinton, A.P.; LoVetri, K.; Yakandawala, N.; Rumbaugh, K.P.; Madhyastha, S. Antibiofilm Efficacy of DispersinB(®) Wound Spray Used in Combination with a Silver Wound Dressing. Microbiol. Insights 2014, 7, 9–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kostenko, V.; Lyczak, J.; Turner, K.; Martinuzzi, R.J. Impact of silver-containing wound dressings on bacterial biofilm viability and susceptibility to antibiotics during prolonged treatment. Antimicrob. Agents Chemother. 2010, 54, 5120–5131. [Google Scholar] [CrossRef] [Green Version]
- Hoekstra, M.J.; Westgate, S.J.; Mueller, S. Povidone-iodine ointment demonstrates in vitro efficacy against biofilm formation. Int. Wound J. 2017, 14, 172–179. [Google Scholar] [CrossRef]
- Parsons, D.; Meredith, K.; Rowlands, V.J.; Short, D.; Metcalf, D.G.; Bowler, P.G. Enhanced Performance and Mode of Action of a Novel Antibiofilm Hydrofiber® Wound Dressing. Biomed. Res. Int. 2016, 2016, 7616471. [Google Scholar] [CrossRef]
- Thorn, R.M.; Greenman, J. A novel in vitro flat-bed perfusion biofilm model for determining the potential antimicrobial efficacy of topical wound treatments. J. Appl. Microbiol. 2009, 107, 2070–2079. [Google Scholar] [CrossRef]
- Percival, S.L.; Bowler, P.; Woods, E.J. Assessing the effect of an antimicrobial wound dressing on biofilms. Wound Repair Regen. 2008, 16, 52–57. [Google Scholar] [CrossRef]
- Kalan, L.R.; Pepin, D.M.; Ul-Haq, I.; Miller, S.B.; Hay, M.E.; Precht, R.J. Targeting biofilms of multidrug-resistant bacteria with silver oxynitrate. Int. J. Antimicrob. Agents 2017, 49, 719–726. [Google Scholar] [CrossRef]
- Bowler, P.G.; Parsons, D. Combatting wound biofilm and recalcitrance with a novel anti-biofilm Hydrofiber® wound dressing. Wound Med. 2016, 14, 6–11. [Google Scholar] [CrossRef] [Green Version]
- Kucera, J.; Sojka, M.; Pavlik, V.; Szuszkiewicz, K.; Velebny, V.; Klein, P. Multispecies biofilm in an artificial wound bed--A novel model for in vitro assessment of solid antimicrobial dressings. J. Microbiol. Methods 2014, 103, 18–24. [Google Scholar] [CrossRef] [PubMed]
- Malone, M.; Goeres, D.M.; Gosbell, I.; Vickery, K.; Jensen, S.; Stoodley, P. Approaches to biofilm-associated infections: The need for standardized and relevant biofilm methods for clinical applications. Expert Rev. Anti-Infect. Ther. 2017, 15, 147–156. [Google Scholar] [CrossRef] [PubMed]
- Thaarup, I.C.; Iversen, A.K.S.; Lichtenberg, M.; Bjarnsholt, T.; Jakobsen, T.H. Biofilm Survival Strategies in Chronic Wounds. Microorganisms 2022, 10, 775. [Google Scholar] [CrossRef]
- Wong, S.Y.; Manikam, R.; Muniandy, S. Prevalence and antibiotic susceptibility of bacteria from acute and chronic wounds in Malaysian subjects. J. Infect. Dev. Ctries. 2015, 9, 936–944. [Google Scholar] [CrossRef] [Green Version]
- Dowd, S.E.; Sun, Y.; Secor, P.R.; Rhoads, D.D.; Wolcott, B.M.; James, G.A.; Wolcott, R.D. Survey of bacterial diversity in chronic wounds using pyrosequencing, DGGE, and full ribosome shotgun sequencing. BMC Microbiol. 2008, 8, 43. [Google Scholar] [CrossRef] [Green Version]
- Dissemond, J.; Schmid, E.N.; Esser, S.; Witthoff, M.; Goos, M. [Bacterial colonization of chronic wounds. Studies on outpatients in a university dermatology clinic with special consideration of ORSA]. Hautarzt 2004, 55, 280–288. [Google Scholar] [CrossRef]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Hobley, L.; Harkins, C.; MacPhee, C.E.; Stanley-Wall, N.R. Giving structure to the biofilm matrix: An overview of individual strategies and emerging common themes. FEMS Microbiol. Rev. 2015, 39, 649–669. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bjarnsholt, T. The role of bacterial biofilms in chronic infections. APMIS 2013, 121, 1–51. [Google Scholar] [CrossRef]
- Roberts, A.E.; Kragh, K.N.; Bjarnsholt, T.; Diggle, S.P. The Limitations of In Vitro Experimentation in Understanding Biofilms and Chronic Infection. J. Mol. Biol. 2015, 427, 3646–3661. [Google Scholar] [CrossRef]
- Werthen, M.; Henriksson, L.; Jensen, P.O.; Sternberg, C.; Givskov, M.; Bjarnsholt, T. An in vitro model of bacterial infections in wounds and other soft tissues. APMIS 2010, 118, 156–164. [Google Scholar] [CrossRef] [PubMed]
- Crone, S.; Garde, C.; Bjarnsholt, T.; Alhede, M. A novel in vitro wound biofilm model used to evaluate low-frequency ultrasonic-assisted wound debridement. J. Wound Care 2015, 24, 64–72. [Google Scholar] [CrossRef]
- Chen, X.; Lorenzen, J.; Xu, Y.; Jonikaite, M.; Thaarup, I.C.; Bjarnsholt, T.; Kirketerp-Møller, K.; Thomsen, T.R. A novel chronic wound biofilm model sustaining coexistence of Pseudomonas aeruginosa and Staphylococcus aureus suitable for testing of antibiofilm effect of antimicrobial solutions and wound dressings. Wound Repair Regen. 2021, 29, 820–829. [Google Scholar] [CrossRef] [PubMed]
- Nedelea, A.G.; Plant, R.L.; Robins, L.I.; Maddocks, S.E. Testing the efficacy of topical antimicrobial treatments using a two- and five-species chronic wound biofilm model. J. Appl. Microbiol. 2022, 132, 715–724. [Google Scholar] [CrossRef] [PubMed]
- DeLeon, S.; Clinton, A.; Fowler, H.; Everett, J.; Horswill, A.R.; Rumbaugh, K.P. Synergistic Interactions of Pseudomonas aeruginosa and Staphylococcus aureus in an In Vitro Wound Model. Infect. Immun. 2014, 82, 4718. [Google Scholar] [CrossRef] [Green Version]
- Woods, J.; Boegli, L.; Kirker, K.R.; Agostinho, A.M.; Durch, A.M.; Delancey Pulcini, E.; Stewart, P.S.; James, G.A. Development and application of a polymicrobial, in vitro, wound biofilm model. J. Appl. Microbiol. 2012, 112, 998–1006. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.; Dowd, S.E.; Smith, E.; Rhoads, D.D.; Wolcott, R.D. In vitro multispecies Lubbock chronic wound biofilm model. Wound Repair Regen. 2008, 16, 805–813. [Google Scholar] [CrossRef]
- Townsend, E.M.; Sherry, L.; Rajendran, R.; Hansom, D.; Butcher, J.; Mackay, W.G.; Williams, C.; Ramage, G. Development and characterisation of a novel three-dimensional inter-kingdom wound biofilm model. Biofouling 2016, 32, 1259–1270. [Google Scholar] [CrossRef] [Green Version]
- Hill, K.E.; Malic, S.; McKee, R.; Rennison, T.; Harding, K.G.; Williams, D.W.; Thomas, D.W. An in vitro model of chronic wound biofilms to test wound dressings and assess antimicrobial susceptibilities. J. Antimicrob. Chemother. 2010, 65, 1195–1206. [Google Scholar] [CrossRef] [Green Version]
- Lipp, C.; Kirker, K.; Agostinho, A.; James, G.; Stewart, P. Testing wound dressings using an in vitro wound model. J. Wound Care 2010, 19, 220–226. [Google Scholar] [CrossRef] [Green Version]
- Junka, A.F.; Żywicka, A.; Szymczyk, P.; Dziadas, M.; Bartoszewicz, M.; Fijałkowski, K.A.D.A.M. test (Antibiofilm Dressing’s Activity Measurement)—Simple method for evaluating anti-biofilm activity of drug-saturated dressings against wound pathogens. J. Microbiol. Methods 2017, 143, 6–12. [Google Scholar] [CrossRef] [PubMed]
- Lenselink, E.; Andriessen, A. A cohort study on the efficacy of a polyhexanide-containing biocellulose dressing in the treatment of biofilms in wounds. J. Wound Care 2011, 20, 534–539. [Google Scholar] [CrossRef] [PubMed]
- Davis, S.C.; Harding, A.; Gil, J.; Parajon, F.; Valdes, J.; Solis, M.; Higa, A. Effectiveness of a polyhexanide irrigation solution on methicillin-resistant Staphylococcus aureus biofilms in a porcine wound model. Int. Wound J. 2017, 14, 937–944. [Google Scholar] [CrossRef] [PubMed]
- Hubner, N.O.; Kramer, A. Review on the efficacy, safety and clinical applications of polihexanide, a modern wound antiseptic. Ski. Pharmacol. Physiol. 2010, 23 (Suppl. S1), 17–27. [Google Scholar] [CrossRef]
- Tahir, S.; Malone, M.; Hu, H.; Deva, A.; Vickery, K. The Effect of Negative Pressure Wound Therapy with and without Instillation on Mature Biofilms In Vitro. Materials 2018, 11, 811. [Google Scholar] [CrossRef] [Green Version]
- Schwarzer, S.; James, G.A.; Goeres, D.; Bjarnsholt, T.; Vickery, K.; Percival, S.L.; Stoodley, P.; Schultz, G.; Jensen, S.O.; Malone, M. The efficacy of topical agents used in wounds for managing chronic biofilm infections: A systematic review. J. Infect. 2020, 80, 261–270. [Google Scholar] [CrossRef]
- Kramer, A.; Dissemond, J.; Kim, S.; Willy, C.; Mayer, D.; Papke, R.; Tuchmann, F.; Assadian, O. Consensus on Wound Antisepsis: Update 2018. Skin Pharmacol. Physiol. 2018, 31, 28–58. [Google Scholar] [CrossRef]
- Stuermer, E.K.; Plattfaut, I.; Dietrich, M.; Brill, F.; Kampe, A.; Wiencke, V.; Ulatowski, A.; Geffken, M.; Rembe, J.D.; Naumova, E.A.; et al. In vitro Activity of Antimicrobial Wound Dressings on P. aeruginosa Wound Biofilm. Front. Microbiol. 2021, 12, 664030. [Google Scholar] [CrossRef]
- Ortega-Peña, S.; Hidalgo-González, C.; Robson, M.C.; Krötzsch, E. In vitro microbicidal, anti-biofilm and cytotoxic effects of different commercial antiseptics. Int. Wound J. 2017, 14, 470–479. [Google Scholar] [CrossRef]
- Vermeulen, H.; van Hattem, J.M.; Storm-Versloot, M.N.; Ubbink, D.T. Topical silver for treating infected wounds. Cochrane Database Syst. Rev. 2007, 1, CD005486. [Google Scholar] [CrossRef]
- Ammons, M.C.; Ward, L.S.; James, G.A. Anti-biofilm efficacy of a lactoferrin/xylitol wound hydrogel used in combination with silver wound dressings. Int. Wound J. 2011, 8, 268–273. [Google Scholar] [CrossRef] [PubMed]
- Percival, S.L.; Bowler, P.G.; Dolman, J. Antimicrobial activity of silver-containing dressings on wound microorganisms using an in vitro biofilm model. Int. Wound J. 2007, 4, 186–191. [Google Scholar] [CrossRef] [PubMed]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reddersen, K.; Tittelbach, J.; Wiegand, C. 3D Biofilm Models Containing Multiple Species for Antimicrobial Testing of Wound Dressings. Microorganisms 2022, 10, 2027. https://doi.org/10.3390/microorganisms10102027
Reddersen K, Tittelbach J, Wiegand C. 3D Biofilm Models Containing Multiple Species for Antimicrobial Testing of Wound Dressings. Microorganisms. 2022; 10(10):2027. https://doi.org/10.3390/microorganisms10102027
Chicago/Turabian StyleReddersen, Kirsten, Jörg Tittelbach, and Cornelia Wiegand. 2022. "3D Biofilm Models Containing Multiple Species for Antimicrobial Testing of Wound Dressings" Microorganisms 10, no. 10: 2027. https://doi.org/10.3390/microorganisms10102027