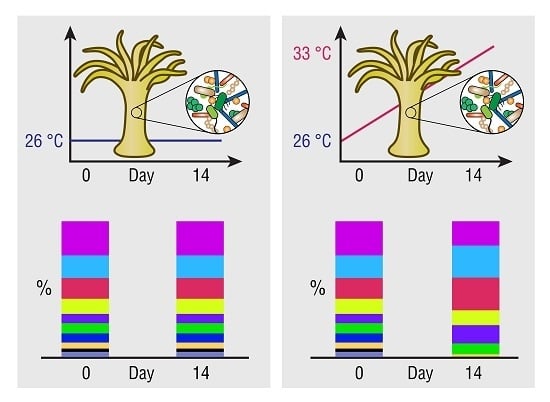
The Effect of Thermal Stress on the Bacterial Microbiome of Exaiptasia diaphana
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Conditions and Sample Processing
2.2. Sequencing Data Workflow
2.3. Physiological and Microbiome Diversity Data Analyses
2.4. Analysis of Changes in Abundance of Selected Bacterial Taxa
2.5. Indicator Species Identification
3. Results
3.1. Sequencing Data and Bacterial Community Characteristics
3.2. Phenotypic Changes in the Anemones
3.3. Changes in Alpha Diversity of the Bacterial Microbiomes
3.4. Changes in Beta Diversity of the Bacterial Microbiomes
3.5. Changes in Abundance of Selected Bacterial Taxa
3.6. Indicator Species Identification
4. Discussion
4.1. Factors Underpinning Bleaching
4.2. Environmental Stressors Reduced Alpha Diversity
4.3. Turnover of Low-Abundance ASVs Drive Shifts in Beta Diversity
4.4. Changes in Bacterial Associates at a High Taxonomic Level were Apparent
4.5. Were Vibrio Victims of Competition
4.6. Specific Bacteria as Biomarkers for Thermal Stress
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Abram, N.J.; McGregor, H.V.; Tierney, J.E.; Evans, M.N.; McKay, N.P.; Kaufman, D.S.; Thirumalai, K.; Martrat, B.; Goosse, H.; Phipps, S.J.; et al. Early onset of industrial-era warming across the oceans and continents. Nature 2016, 536, 411–417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coles, S.L.; Jokiel, P.L.; Lewis, C.R. Thermal tolerance in tropical versus subtropical Pacific reef corals. Pac. Sci. 1976, 30, 159–166. [Google Scholar]
- Hughes, T.P.; Kerry, J.T.; Álvarez-Noriega, M.; Álvarez-Romero, J.G.; Anderson, K.D.; Baird, A.H.; Babcock, R.C.; Beger, M.; Bellwood, D.R.; Berkelmans, R.; et al. Global warming and recurrent mass bleaching of corals. Nature 2017, 543, 373–377. [Google Scholar] [CrossRef]
- Tremblay, P.; Grover, R.; Maguer, J.; Legendre, L.; Ferrier-Pages, C. Autotrophic carbon budget in coral tissue: A new C-13-based model of photosynthate translocation. J. Exp. Biol. 2012, 215, 1384–1393. [Google Scholar] [CrossRef] [Green Version]
- Ostrander, G.K.; Armstrong, K.M.; Knobbe, E.T.; Gerace, D.; Scully, E.P. Rapid transition in the structure of a coral reef community: The effects of coral bleaching and physical disturbance. Proc. Natl. Acad. Sci. USA 2000, 97, 5297–5302. [Google Scholar] [CrossRef] [Green Version]
- Rohwer, F.; Seguritan, V.; Azam, F.; Knowlton, N. Diversity and distribution of coral-associated bacteria. Mar. Ecol. Prog. Ser. 2002, 243, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Krediet, C.J.; Ritchie, K.B.; Paul, V.J.; Teplitski, M. Coral-associated micro-organisms and their roles in promoting coral health and thwarting diseases. Proc. R. Soc. 2013, 280. [Google Scholar] [CrossRef] [Green Version]
- Sharp, K.H.; Ritchie, K.B. Multi-partner interactions in corals in the face of climate change. Biol. Bull. 2012, 223, 66–77. [Google Scholar] [CrossRef] [Green Version]
- Cai, L.; Zhou, G.; Tong, H.; Tian, R.-M.; Zhang, W.; Ding, W.; Liu, S.; Huang, H.; Qian, P.-Y. Season structures prokaryotic partners but not algal symbionts in subtropical hard corals. Appl. Microbiol. Biotechnol. 2018, 102, 4963–4973. [Google Scholar] [CrossRef]
- Sharp, K.H.; Pratte, Z.A.; Kerwin, A.H.; Rotjan, R.D.; Stewart, F.J. Season, but not symbiont state, drives microbiome structure in the temperate coral Astrangia poculata. Microbiome 2017, 5, 120. [Google Scholar] [CrossRef]
- Ziegler, M.; Seneca, F.O.; Yum, L.K.; Palumbi, S.R.; Voolstra, C.R. Bacterial community dynamics are linked to patterns of coral heat tolerance. Nat. Commun. 2017, 8, 14213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.T.M.; Davy, S.K.; Tang, S.-L.; Kench, P.S. Mucus sugar content shapes the bacterial community structure in thermally stressed Acropora muricata. Front. Microbiol. 2016, 7, 371. [Google Scholar] [CrossRef] [PubMed]
- Tracy, A.M.; Koren, O.; Douglas, N.; Weil, E.; Harvell, C.D. Persistent shifts in Caribbean coral microbiota are linked to the 2010 warm thermal anomaly. Environ. Microbiol. Rep. 2015, 7, 471–479. [Google Scholar] [CrossRef] [PubMed]
- Gajigan, A.P.; Diaz, L.A.; Conaco, C. Resilience of the prokaryotic microbial community of Acropora digitifera to elevated temperature. MicrobiologyOpen 2017, 6, e00478. [Google Scholar] [CrossRef] [PubMed]
- Bourne, D.G.; Garren, M.; Work, T.M.; Rosenberg, E.; Smith, G.W.; Harvell, C.D. Microbial disease and the coral holobiont. Trends Microbiol. 2009, 17, 554–562. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.R.; Rivera, H.E.; Closek, C.J.; Medina, M. Microbes in the coral holobiont: Partners through evolution, development, and ecological interactions. Front. Cell. Infect. Microbiol. 2015, 4, 176. [Google Scholar] [CrossRef] [PubMed]
- Mouchka, M.E.; Hewson, I.; Harvell, C.D. Coral-associated bacterial assemblages: Current knowledge and the potential for climate-driven impacts. Integr. Comp. Biol. 2010, 50, 662–674. [Google Scholar] [CrossRef]
- Weis, V.M.; Davy, S.K.; Hoegh-Guldberg, O.; Rodriguez-Lanetty, M.; Pringle, J.R. Cell biology in model systems as the key to understanding corals. Trends Ecol. Evol. 2008, 23, 369–376. [Google Scholar] [CrossRef]
- Voolstra, C.R. A journey into the wild of the cnidarian model system Aiptasia and its symbionts. Mol. Ecol. 2013, 22, 4366–4368. [Google Scholar] [CrossRef]
- Tolleter, D.; Seneca, F.O.; DeNofrio, J.C.; Krediet, C.J.; Palumbi, S.R.; Pringle, J.R.; Grossman, A.R. Coral bleaching independent of photosynthetic activity. Curr. Biol. 2013, 23, 1782–1786. [Google Scholar] [CrossRef] [Green Version]
- Bieri, T.; Onishi, M.; Xiang, T.; Grossman, A.R.; Pringle, J.R. Relative contributions of various cellular mechanisms to loss of algae during cnidarian bleaching. PLoS ONE 2016, 11, e0152693. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gates, R.D.; Baghdasarian, G.; Muscatine, L. Temperature stress causes host-cell detachment in symbiotic cnidarians: Implications for coral bleaching. Biol. Bull. 1992, 182, 324–332. [Google Scholar] [CrossRef] [PubMed]
- Hillyer, K.E.; Tumanov, S.; Villas-Bôas, S.; Davy, S.K. Metabolite profiling of symbiont and host during thermal stress and bleaching in a model cnidarian–dinoflagellate symbiosis. J. Exp. Biol. 2016, 219, 516–527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Núñez-Pons, L.; Bertocci, I.; Baghdasarian, G. Symbiont dynamics during thermal acclimation using cnidarian-dinoflagellate model holobionts. Mar. Environ. Res. 2017, 130, 303–314. [Google Scholar] [CrossRef] [PubMed]
- Gegner, H.M.; Ziegler, M.; Rädecker, N.; Buitrago-López, C.; Aranda, M.; Voolstra, C.R. High salinity conveys thermotolerance in the coral model Aiptasia. Biol. Open 2017, 6, 1943–1948. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Plovie, A. Comparison of Bacterial Communities Associated with Healthy and Bleached Aiptasia pallida, a Novel Model Organism for Coral Studies: Implications and Variation during Bleaching. Biochemistry and Biotechnology. Master’s Thesis, Ghent University, Ghent, Belgium, 2010. [Google Scholar]
- Ahmed, H.I.; Herrera, M.; Liew, Y.J.; Aranda, M. Long-term temperature stress in the coral model Aiptasia supports the “Anna Karenina Principle” for bacterial microbiomes. Front. Microbiol. 2019, 10, 975. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zaneveld, J.R.; McMinds, R.; Vega Thurber, R. Stress and stability: Applying the Anna Karenina principle to animal microbiomes. Nat. Microbiol. 2017, 2, 17121. [Google Scholar] [CrossRef]
- Dungan, A.M.; Hartman, L.M.; Tortorelli, G.; Belderock, R.; Lamb, A.M.; Pisan, L.; McFadden, G.; Blackall, L.L.; van Oppen, M.J.H. Exaiptasia diaphana from the Great Barrier Reef: A valuable resource for coral symbiosis research. bioRxiv 2019. [Google Scholar] [CrossRef] [Green Version]
- Röthig, T.; Costa, R.M.; Simona, F.; Baumgarten, S.; Torres, A.F.; Radhakrishnan, A.; Aranda, M.; Voolstra, C.R. Distinct bacterial communities associated with the coral model Aiptasia in aposymbiotic and symbiotic states with Symbiodinium. Front. Mar. Sci. 2016, 3, 234. [Google Scholar] [CrossRef] [Green Version]
- Marty-Rivera, M.; Yudowski, G.; Roberson, L. Mitigation of coral bleaching by antioxidants. bioRxiv 2018. [Google Scholar] [CrossRef]
- Zaragoza, W.J.; Krediet, C.J.; Meyer, J.L.; Canas, G.; Ritchie, K.B.; Teplitski, M. Outcomes of infections of sea anemone Aiptasia pallida with Vibrio spp. pathogenic to corals. Microb. Ecol. 2014, 68, 388–396. [Google Scholar] [CrossRef] [PubMed]
- Warner, M.E.; Fitt, W.K.; Schmidt, G.W. Damage to photosystem II in symbiotic dinoflagellates: A determinant of coral bleaching. Proc. Natl. Acad. Sci. USA 1999, 96, 8007–8012. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Wilson, K.; Li, Y.; Whan, V.; Lehnert, S.; Byrne, K.; Moore, S.; Pongsomboon, S.; Tassanakajon, A.; Rosenberg, G.; Ballment, E.; et al. Genetic mapping of the black tiger shrimp Penaeus monodon with amplified fragment length polymorphism. Aquaculture 2002, 204, 297–309. [Google Scholar] [CrossRef]
- Andersson, A.F.; Lindberg, M.; Jakobsson, H.; Bäckhed, F.; Nyrén, P.; Engstrand, L. Comparative analysis of human gut microbiota by barcoded pyrosequencing. PLoS ONE 2008, 3, e2836. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Garrith, G.M.; Tiedje, J.M.; Cole, J.R. Naive bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [Green Version]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Caporaso, J.G. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2′s q2-feature-classifier plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef] [PubMed]
- R.C. Team. R: A Language and Environment for Statisitical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018. [Google Scholar]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; Stevens, M.H.H.; Wagner, H. Vegan: Community Ecology Package, R package version 2.5–6; Available online: https://cran.r-project.org/src/contrib/vegan_2.5-6.tar.gz (accessed on 20 December 2019).
- Davis, N.M.; Proctor, D.M.; Holmes, S.P.; Relman, D.A.; Callahan, B.J. Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome 2018, 6, 226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis, R package version 3.2.1 ed.; Springer: New York, NY, USA, 2016. [Google Scholar]
- Shapiro, S.S.; Wilk, M.B. An analysis of variance test for normality complete samples. Biometrika 1965, 52, 591–611. [Google Scholar] [CrossRef]
- Levene, H. Robust tests for equality of variances. In Contributions to Probability and Statistics: Essays in Honor of Harold Hotelling; Olkin, I., Ghurye, S.G., Hoeffding, W., Madow, W.G., Mann, H.B., Eds.; Stanford University Press: Menlo Park, CA, USA, 1960; pp. 278–292. [Google Scholar]
- Fox, J.; Weisberg, S.; Price, B. Car: Companion to Applied Regression, R package version 3.0–5; Available online: https://cran.r-project.org/src/contrib/car_3.0-5.tar.gz (accessed on 20 December 2019).
- Student. The probable error of a mean. Biometrika 1908, 6, 1–25. [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Kruskal, W.H.; Wallis, W.A. Use of ranks in one-criterion variance analysis. J. Am. Stat. Assoc. 1952, 47, 583–621. [Google Scholar] [CrossRef]
- Welch, B.L. The generalization of ‘Student’s’ problem when several different population variances are involved. Biometrika 1947, 34, 28–35. [Google Scholar] [CrossRef]
- Whitney, J. Testing for differences with the nonparametric Mann–Whitney U test. J. Wound Ostomy Cont. Nurs. 1997, 24, 12. [Google Scholar] [CrossRef]
- Simpson, E.H. Measurement of diversity. Nature 1949, 163, 688. [Google Scholar] [CrossRef]
- Shannon, C.E.; Weaver, W. The Mathematical Theory of Communication; University of Illinois Press: Champaign, IL, USA, 1949. [Google Scholar]
- Warnes, G.R.; Bolker, B.; Bonebakker, L.; Gentleman, R.; Huber, W.; Liaw, A.; Lumley, T.; Maechler, M.; Magnusson, A.; Moeller, S.; et al. Gplots: Various R Programming Tools for Plotting Data, R package version 3.0.1.1; Available online: https://cran.r-project.org/src/contrib/gplots_3.0.1.1.tar.gz (accessed on 20 December 2019).
- Wang, Y.; Naumann, U.; Wright, S.T.; Warton, D.I. Mvabund—An R package for model-based analysis of multivariate abundance. Methods Ecol. Evol. 2012, 3, 471–474. [Google Scholar] [CrossRef]
- Kimes, N.E.; Grim, C.J.; Johnson, W.R.; Hasan, N.A.; Tall, B.D.; Kothary, M.H.; Kiss, H.; Munk, A.C.; Tapia, R.; Green, L.; et al. Temperature regulation of virulence factors in the pathogen Vibrio Coralliilyticus. ISME J. 2012, 6, 835–846. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roberts, D.W. labdsv: Ordination and Multivariate Analysis for Ecology, R package version 1.8.0; 2016. Available online: https://cran.r-project.org/web/packages/labdsv/labdsv.pdf (accessed on 20 December 2019).
- Glasl, B.; Webster, N.S.; Bourne, D.G. Microbial indicators as a diagnostic tool for assessing water quality and climate stress in coral reef ecosystems. Mar. Biol. 2017, 164, 91. [Google Scholar] [CrossRef]
- Glasl, B.; Herndl, G.J.; Frade, P.R. The microbiome of coral surface mucus has a key role in mediating holobiont health and survival upon disturbance. ISME J. 2016, 10, 2280–2292. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Chen, Q.; Long, L.-J.; Dong, J.-D.; Yang, J.; Zhang, S. Bacterial dynamics within the mucus, tissue and skeleton of the coral Porites lutea during different seasons. Sci. Rep. 2014, 4, 7320. [Google Scholar] [CrossRef] [Green Version]
- Astudillo-García, C.; Bell, J.J.; Webster, N.S.; Glasl, B.; Jompa, J.; Montoya, J.M.; Taylor, M.W. Evaluating the core microbiota in complex communities: A systematic investigation. Environ. Microbiol. 2017, 19, 1450–1462. [Google Scholar] [CrossRef]
- Dufrêne, M.; Legendre, P. Species assemblages and indicator species: The need for a flexible asymmetrical approach. Ecol. Monogr. 1997, 67, 345–366. [Google Scholar] [CrossRef]
- Clayton, W.S.; Lasker, H.R. Host feeding regime and Zooxanthellal photosynthesis in the anemone, Aiptasia pallida (Verrill). Biol. Bull. 1984, 167, 590–600. [Google Scholar] [CrossRef]
- Cook, C.B.; D’Elia, C.F.; Muller-Parker, G. Host feeding and nutrient sufficiency for zooxanthellae in the sea anemone Aiptasia pallida. Mar. Biol. 1988, 98, 253–262. [Google Scholar] [CrossRef]
- Davy, S.; Cook, C. The relationship between nutritional status and carbon flux in the zooxanthellate sea anemone Aiptasia pallida. Mar. Biol. 2001, 139, 999–1005. [Google Scholar] [CrossRef]
- Lehnert, E.M.; Mouchka, M.E.; Burriesci, M.S.; Gallo, N.D.; Schwarz, J.A.; Pringle, J.R. Extensive differences in gene expression between symbiotic and aposymbiotic cnidarians. G3 2014, 4, 277–295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grottoli, A.G.; Rodrigues, L.J.; Palardy, J.E. Heterotrophic plasticity and resilience in bleached corals. Nature 2006, 440, 1186–1189. [Google Scholar] [CrossRef] [PubMed]
- Borell, E.M.; Yuliantri, A.R.; Bischof, K.; Richter, C. The effect of heterotrophy on photosynthesis and tissue composition of two scleractinian corals under elevated temperature. J. Exp. Mar. Biol. Ecol. 2008, 364, 116–123. [Google Scholar] [CrossRef]
- Ferrier-Pagès, C.; Rottier, C.; Beraud, E.; Levy, O. Experimental assessment of the feeding effort of three scleractinian coral species during a thermal stress: Effect on the rates of photosynthesis. J. Exp. Mar. Biol. Ecol. 2010, 390, 118–124. [Google Scholar] [CrossRef]
- Grottoli, A.G.; Dalcin Martins, P.; Wilkins, M.J.; Johnston, M.D.; Warner, M.E.; Cai, W.-J.; Melman, T.F.; Hoadley, K.D.; Pettay, D.T.; Levas, S.; et al. Coral physiology and microbiome dynamics under combined warming and ocean acidification. PLoS ONE 2018, 13, e0191156. [Google Scholar] [CrossRef]
- Rocca, J.D.; Simonin, M.; Blaszczak, J.R.; Ernakovich, J.G.; Gibbons, S.M.; Midani, F.S.; Washburne, A.D. The Microbiome Stress Project: Toward a global meta-analysis of environmental stressors and their effects on microbial communities. Front. Microbiol. 2019, 9, 3272. [Google Scholar] [CrossRef]
- Bourne, D.; Iida, Y.; Uthicke, S.; Smith-Keune, C. Changes in coral-associated microbial communities during a bleaching event. ISME J. 2008, 2, 350–363. [Google Scholar] [CrossRef]
- Santos, H.F.; Carmo, F.L.; Duarte, G.; Dini-Andreote, F.; Castro, C.B.; Rosado, A.S.; van Elsas, J.D.; Peixoto, R.S. Climate change affects key nitrogen-fixing bacterial populations on coral reefs. ISME J. 2014, 8, 2272–2279. [Google Scholar] [CrossRef] [Green Version]
- Tout, J.; Siboni, N.; Messer, L.F.; Garren, M.; Stocker, R.; Webster, N.S.; Ralph, P.J.; Seymour, J.R. Increased seawater temperature increases the abundance and alters the structure of natural Vibrio populations associated with the coral Pocillopora damicornis. Front. Microbiol. 2015, 6, 432. [Google Scholar] [CrossRef] [Green Version]
- Pratte, Z.A.; Richardson, L.L.; Mills, D.K. Microbiota shifts in the surface mucopolysaccharide layer of corals transferred from natural to aquaria settings. J. Invertebr. Pathol. 2015, 125, 42–44. [Google Scholar] [CrossRef]
- Hester, E.R.; Barott, K.L.; Nulton, J.; Vermeij, M.J.A.; Rohwer, F.L. Stable and sporadic symbiotic communities of coral and algal holobionts. ISME J. 2016, 10, 1157–1169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sweet, M.J.; Brown, B.E.; Dunne, R.P.; Singleton, I.; Bulling, M. Evidence for rapid, tide-related shifts in the microbiome of the Coral Coelastrea Aspera. Coral Reefs 2017, 36, 815–828. [Google Scholar] [CrossRef] [Green Version]
- Brown, T.; Otero, C.; Grajales, A.; Rodríguez, E.; Rodriguez-Lanetty, M. Worldwide exploration of the microbiome harbored by the cnidarian model, Exaiptasia pallida (Agassiz in Verrill, 1864) indicates a lack of bacterial association specificity at a lower taxonomic rank. PeerJ 2017, 5, e3235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reshef, L.; Koren, O.; Loya, Y.; Zilber-Rosenberg, I.; Rosenberg, E. The coral probiotic hypothesis. Environ. Microbiol. 2006, 8, 2068–2073. [Google Scholar] [CrossRef] [Green Version]
- Shade, A.; Jones, S.E.; Caporaso, J.G.; Handelsman, J.; Knight, R.; Fierer, N.; Gilbert, J.A. Conditionally rare taxa disproportionately contribute to temporal changes in microbial diversity. mBio 2014, 5, e01371-14. [Google Scholar] [CrossRef] [Green Version]
- Vezzulli, L.; Previati, M.; Pruzzo, C.; Marchese, A.; Bourne, D.G.; Cerrano, C. Vibrio infections triggering mass mortality events in a warming Mediterranean Sea. Environ. Microbiol. 2010, 12, 2007–2019. [Google Scholar] [CrossRef]
- Munn, C.B. The role of Vibrios in diseases of corals. Microbiol. Spectr. 2015, 3. [Google Scholar] [CrossRef] [Green Version]
- Welsh, R.M.; Zaneveld, J.R.; Rosales, S.M.; Payet, J.P.; Burkepile, D.E.; Thurber, R.V. Bacterial predation in a marine host-associated microbiome. ISME J. 2015, 10, 1540–1544. [Google Scholar] [CrossRef]
- Ruiz-Ponte, C.; Samain, J.F.; Sánchez, J.L.; Nicolas, J.L. The benefit of a Roseobacter species on the survival of scallop larvae. Mar. Biotechnol. 1999, 1, 52–59. [Google Scholar] [CrossRef]
- Hibbing, M.E.; Fuqua, C.; Parsek, M.R.; Peterson, S.B. Bacterial competition: Surviving and thriving in the microbial jungle. Nat. Rev. Microbiol. 2010, 8, 15–25. [Google Scholar] [CrossRef] [Green Version]
- Furusawa, G.; Yoshikawa, T.; Yasuda, A.; Sakata, T. Algicidal activity and gliding motility of Saprospira sp. SS98-5. Can. J. Microbiol. Rev. Can. Microbiol. 2003, 49, 92–100. [Google Scholar] [CrossRef] [PubMed]
- Shi, M.; Zou, M.; Liu, X.; Gao, Y.; Zhang, Z.; Wu, W.; Wen, D.; Chen, Z.; An, C. A novel bacterium Saprospira sp. strain PdY3 forms bundles and lyses cyanobacteria. Front. Biosci. 2006, 11, 1916–1923. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lewin, R.A. Saprospira grandis: A flexibacterium that can catch bacterial prey by ixotrophy. Microb. Ecol. 1997, 34, 232–236. [Google Scholar] [CrossRef] [PubMed]
- Weiler, B.A.; Verhoeven, J.T.P.; Dufour, S.C. Bacterial communities in tissues and surficial mucus of the cold-water coral Paragorgia Arborea. Front. Mar. Sci. 2018, 5. [Google Scholar] [CrossRef]
- Hoegh-Guldberg, O.; Smith, G.J. Influence of the population density of zooxanthellae and supply of ammonium on the biomass and metabolic characteristics of the reef corals Seriatopora hystrix and Stylophora pistillata. Mar. Ecol. Prog. Ser. 1989, 57, 173–186. [Google Scholar] [CrossRef]
- Muscatine, L.; Falkowski, P.G.; Dubinsky, Z.; Cook, P.A.; McCloskey, L.R.; Smith, D.C. The effect of external nutrient resources on the population dynamics of zooxanthellae in a reef coral. Proc. R. Soc. B 1989, 236, 311–324. [Google Scholar] [CrossRef]
- Kimes, N.E.; Johnson, W.R.; Torralba, M.; Nelson, K.E.; Weil, E.; Morris, P.J. The Montastraea faveolata microbiome: Ecological and temporal influences on a Caribbean reef-building coral in decline. Environ. Microbiol. 2013, 15, 2082–2094. [Google Scholar] [CrossRef]
- Lawler, S.N.; Kellogg, C.A.; France, S.C.; Clostio, R.W.; Brooke, S.D.; Ross, S.W. Coral-associated bacterial diversity is conserved across two deep-sea Anthothela species. Front. Microbiol. 2016, 7, 458. [Google Scholar] [CrossRef]
- Closek, C.J.; Sunagawa, S.; DeSalvo, M.K.; Piceno, Y.M.; DeSantis, T.Z.; Brodie, E.L.; Weber, M.X.; Voolstra, C.R.; Andersen, G.L.; Medina, M. Coral transcriptome and bacterial community profiles reveal distinct Yellow Band Disease states in Orbicella faveolata. ISME J. 2014, 8, 2411. [Google Scholar] [CrossRef] [Green Version]
- Hollants, J.; Leliaert, F.; Verbruggen, H.; Willems, A.; De Clerck, O. Permanent residents or temporary lodgers: Characterizing intracellular bacterial communities in the siphonous green alga Bryopsis. Proc. R. Soc. B 2013, 280. [Google Scholar] [CrossRef] [Green Version]
- Schwenk, D.; Nohynek, L.; Rischer, H. Algae–bacteria association inferred by 16S rDNA similarity in established microalgae cultures. MicrobiologyOpen 2014, 3, 356–368. [Google Scholar] [CrossRef] [PubMed]
- Jackson, D.A. Compositional data in community ecology: The paradigm or peril of proportions? Ecology 1997, 78, 929–941. [Google Scholar] [CrossRef]
- Vandeputte, D.; Kathagen, G.; D’hoe, K.; Vieira-Silva, S.; Valles-Colomer, M.; Sabino, J.; Wang, J.; Tito, R.Y.; De Commer, L.; Darzi, Y.; et al. Quantitative microbiome profiling links gut community variation to microbial load. Nature 2017, 551, 507–511. [Google Scholar] [CrossRef] [PubMed]
- Props, R.; Kerckhof, F.M.; Rubbens, P.; De Vrieze, J.; Hernandez Sanabria, E.; Waegeman, W.; Monsieurs, P.; Hammes, F.; Boon, N. Absolute quantification of microbial taxon abundances. ISME J. 2017, 11, 584–587. [Google Scholar] [CrossRef] [Green Version]
- Tourlousse, D.M.; Yoshiike, S.; Ohashi, A.; Matsukura, S.; Noda, N.; Sekiguchi, Y. Synthetic spike-in standards for high-throughput 16S rRNA gene amplicon sequencing. Nucleic Acids Res. 2017, 45, e23. [Google Scholar] [CrossRef] [Green Version]
- Stämmler, F.; Gläsner, J.; Hiergeist, A.; Holler, E.; Weber, D.; Oefner, P.J.; Gessner, A.; Spang, R. Adjusting microbiome profiles for differences in microbial load by spike-in bacteria. Microbiome 2016, 4, 28. [Google Scholar] [CrossRef] [Green Version]








© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hartman, L.M.; van Oppen, M.J.H.; Blackall, L.L. The Effect of Thermal Stress on the Bacterial Microbiome of Exaiptasia diaphana. Microorganisms 2020, 8, 20. https://doi.org/10.3390/microorganisms8010020
Hartman LM, van Oppen MJH, Blackall LL. The Effect of Thermal Stress on the Bacterial Microbiome of Exaiptasia diaphana. Microorganisms. 2020; 8(1):20. https://doi.org/10.3390/microorganisms8010020
Chicago/Turabian StyleHartman, Leon M., Madeleine J. H. van Oppen, and Linda L. Blackall. 2020. "The Effect of Thermal Stress on the Bacterial Microbiome of Exaiptasia diaphana" Microorganisms 8, no. 1: 20. https://doi.org/10.3390/microorganisms8010020