YycH and YycI Regulate Expression of Staphylococcus aureus Autolysins by Activation of WalRK Phosphorylation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Creation of yycH, yycI, and yycHI Antisense Knockdown Clones and yycHI Deletion
2.2. Western Blot for Detection of YycH and YycI in Cell Lysates
2.3. Detection of WalR Phosphorylation by Phos-Tag SDS-PAGE and Western Blot
2.4. Triton X-100 Autolysis and Light Microscopy
2.5. WTA Purification and Quantification
2.6. Peptidoglycan Purification and Muropeptide Analysis of the Antisense yycHI Strain
2.7. Zymogram Analysis of Autolysin Extracts
2.8. Overexpression and Purification of WalK, YycH, and YycI
2.9. Reconstitution of Membrane Proteins into Phospholipid Liposomes
2.10. Phosphorylation Assays with WalK in Phospholipid Liposomes
2.11. Phosphorylation Assays with Peptidoglycan Fragments, D-Alanine, and WTA in Detergent Micelles
3. Results
3.1. Inactivation of yycHI Expression Leads to Reduced Autolysis
3.2. Reduction in YycH and YycI Levels Increases WTA Content of S. aureus Cell Walls
3.3. Muropeptide Composition of the Cell Wall Is Not Affected by YycH and YycI
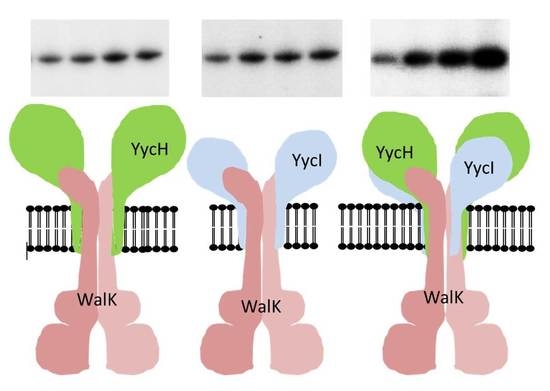
3.4. Both Accessory Proteins YycH and YycI Are Required for Full Activation of WalK Autophosphorylation in Phospholipid Liposomes
3.5. YycH and YycI Stimulate WalR Phosphorylation
3.6. Cell Wall Fragments Did Not Affect WalRK Phosphorylation in In Vitro Phosphorylation Assays
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Villanueva, M.; García, B.; Valle, J.; Rapún, B.; Ruiz de Los Mozos, I.; Solano, C.; Martí, M.; Penadés, J.R.; Toledo-Arana, A.; Lasa, I. Sensory deprivation in Staphylococcus aureus. Nat. Commun. 2018, 9, 523. [Google Scholar] [CrossRef] [PubMed]
- Dubrac, S.; Msadek, T. Identification of Genes Controlled by the Essential YycG/YycF Two-Component System of Staphylococcus aureus. J. Bacteriol. 2004, 186, 1175–1181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dubrac, S.; Boneca, I.G.; Poupel, O.; Msadek, T. New insights into the WalK/WalR (YycG/YycF) essential signal transduction pathway reveal a major role in controlling cell wall metabolism and biofilm formation in Staphylococcus aureus. J. Bacteriol. 2007, 189, 8257–8269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delauné, A.; Dubrac, S.; Blanchet, C.; Poupel, O.; Mäder, U.; Hiron, A.; Leduc, A.; Fitting, C.; Nicolas, P.; Cavaillon, J.-M.; et al. The WalKR system controls major staphylococcal virulence genes and is involved in triggering the host inflammatory response. Infect. Immun. 2012, 80, 3438–3453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fabret, C.; Hoch, J.A. A two-component signal transduction system essential for growth of Bacillus subtilis: Implications for anti-infective therapy. J. Bacteriol. 1998, 180, 6375–6383. [Google Scholar] [CrossRef]
- Martin, P.K.; Li, T.; Sun, D.; Biek, D.P.; Schmid, M.B. Role in cell permeability of an essential two-component system in Staphylococcus aureus. J. Bacteriol. 1999, 181, 3666–3673. [Google Scholar] [CrossRef] [Green Version]
- Poupel, O.; Moyat, M.; Groizeleau, J.; Antunes, L.C.S.; Gribaldo, S.; Msadek, T.; Dubrac, S. Transcriptional Analysis and Subcellular Protein Localization Reveal Specific Features of the Essential WalKR System in Staphylococcus aureus. PLoS ONE 2016, 11, e0151449. [Google Scholar] [CrossRef]
- Burian, M.; Rautenberg, M.; Kohler, T.; Fritz, M.; Krismer, B.; Unger, C.; Hoffmann, W.H.; Peschel, A.; Wolz, C.; Goerke, C. Temporal expression of adhesion factors and activity of global regulators during establishment of Staphylococcus aureus nasal colonization. J. Infect. Dis. 2010, 201, 1414–1421. [Google Scholar] [CrossRef] [Green Version]
- Monk, I.R.; Shaikh, N.; Begg, S.L.; Gajdiss, M.; Sharkey, L.K.R.; Lee, J.Y.H.; Pidot, S.J.; Seemann, T.; Kuiper, M.; Winnen, B.; et al. Zinc-binding to the cytoplasmic PAS domain regulates the essential WalK histidine kinase of Staphylococcus aureus. Nat. Commun. 2019, 10, 3067. [Google Scholar] [CrossRef] [Green Version]
- Jansen, A.; Türck, M.; Szekat, C.; Nagel, M.; Clever, I.; Bierbaum, G. Role of insertion elements and yycFG in the development of decreased susceptibility to vancomycin in Staphylococcus aureus. Int. J. Med. Microbiol. 2007, 297, 205–215. [Google Scholar] [CrossRef]
- Howden, B.P.; McEvoy, C.R.E.; Allen, D.L.; Chua, K.; Gao, W.; Harrison, P.F.; Bell, J.; Coombs, G.; Bennett-Wood, V.; Porter, J.L.; et al. Evolution of multidrug resistance during Staphylococcus aureus infection involves mutation of the essential two component regulator WalKR. PLoS Pathog. 2011, 7, e1002359. [Google Scholar] [CrossRef]
- Gardete, S.; Kim, C.; Hartmann, B.M.; Mwangi, M.; Roux, C.M.; Dunman, P.M.; Chambers, H.F.; Tomasz, A. Genetic pathway in acquisition and loss of vancomycin resistance in a methicillin resistant Staphylococcus aureus (MRSA) strain of clonal type USA300. PLoS Pathog. 2012, 8, e1002505. [Google Scholar] [CrossRef] [Green Version]
- Cameron, D.R.; Jiang, J.-H.; Kostoulias, X.; Foxwell, D.J.; Peleg, A.Y. Vancomycin susceptibility in methicillin-resistant Staphylococcus aureus is mediated by YycHI activation of the WalRK essential two-component regulatory system. Sci. Rep. 2016, 6, 30823. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.; Murakami, H.; Kuwahara-Arai, K.; Hanaki, H.; Hiramatsu, K. Contribution of a thickened cell wall and its glutamine nonamidated component to the vancomycin resistance expressed by Staphylococcus aureus Mu50. Antimicrob. Agents Chemother. 2000, 44, 2276–2285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pereira, P.M.; Filipe, S.R.; Tomasz, A.; Pinho, M.G. Fluorescence ratio imaging microscopy shows decreased access of vancomycin to cell wall synthetic sites in vancomycin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2007, 51, 3627–3633. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szurmant, H.; Mohan, M.A.; Imus, P.M.; Hoch, J.A. YycH and YycI interact to regulate the essential YycFG two-component system in Bacillus subtilis. J. Bacteriol. 2007, 189, 3280–3289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szurmant, H.; Zhao, H.; Mohan, M.A.; Hoch, J.A.; Varughese, K.I. The crystal structure of YycH involved in the regulation of the essential YycFG two-component system in Bacillus subtilis reveals a novel tertiary structure. Protein Sci. 2006, 15, 929–934. [Google Scholar] [CrossRef] [Green Version]
- Santelli, E.; Liddington, R.C.; Mohan, M.A.; Hoch, J.A.; Szurmant, H. The crystal structure of Bacillus subtilis YycI reveals a common fold for two members of an unusual class of sensor histidine kinase regulatory proteins. J. Bacteriol. 2007, 189, 3290–3295. [Google Scholar] [CrossRef] [Green Version]
- Szurmant, H.; Nelson, K.; Kim, E.-J.; Perego, M.; Hoch, J.A. YycH regulates the activity of the essential YycFG two-component system in Bacillus subtilis. J. Bacteriol. 2005, 187, 5419–5426. [Google Scholar] [CrossRef] [Green Version]
- Dobihal, G.S.; Brunet, Y.R.; Flores-Kim, J.; Rudner, D.Z. Homeostatic control of cell wall hydrolysis by the WalRK two-component signaling pathway in Bacillus subtilis. Elife 2019, 8. [Google Scholar] [CrossRef]
- Szurmant, H.; Bu, L.; Brooks, C.L.; Hoch, J.A. An essential sensor histidine kinase controlled by transmembrane helix interactions with its auxiliary proteins. Proc. Natl. Acad. Sci. USA 2008, 105, 5891–5896. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fukushima, T.; Furihata, I.; Emmins, R.; Daniel, R.A.; Hoch, J.A.; Szurmant, H. A role for the essential YycG sensor histidine kinase in sensing cell division. Mol. Microbiol. 2011, 79, 503–522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fukushima, T.; Szurmant, H.; Kim, E.-J.; Perego, M.; Hoch, J.A. A sensor histidine kinase co-ordinates cell wall architecture with cell division in Bacillus subtilis. Mol. Microbiol. 2008, 69, 621–632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mwangi, M.M.; Wu, S.W.; Zhou, Y.; Sieradzki, K.; de Lencastre, H.; Richardson, P.; Bruce, D.; Rubin, E.; Myers, E.; Siggia, E.D.; et al. Tracking the in vivo evolution of multidrug resistance in Staphylococcus aureus by whole-genome sequencing. Proc. Natl. Acad. Sci. USA 2007, 104, 9451–9456. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Forsyth, R.A.; Haselbeck, R.J.; Ohlsen, K.L.; Yamamoto, R.T.; Xu, H.; Trawick, J.D.; Wall, D.; Wang, L.; Brown-Driver, V.; Froelich, J.M.; et al. A genome-wide strategy for the identification of essential genes in Staphylococcus aureus. Mol. Microbiol. 2002, 43, 1387–1400. [Google Scholar] [CrossRef]
- Herbert, S.; Ziebandt, A.-K.; Ohlsen, K.; Schäfer, T.; Hecker, M.; Albrecht, D.; Novick, R.; Götz, F. Repair of global regulators in Staphylococcus aureus 8325 and comparative analysis with other clinical isolates. Infect. Immun. 2010, 78, 2877–2889. [Google Scholar] [CrossRef] [Green Version]
- Monk, I.R.; Tree, J.J.; Howden, B.P.; Stinear, T.P.; Foster, T.J. Complete Bypass of Restriction Systems for Major Staphylococcus aureus Lineages. MBio 2015, 6, e00308-15. [Google Scholar] [CrossRef] [Green Version]
- Schlag, M.; Biswas, R.; Krismer, B.; Kohler, T.; Zoll, S.; Yu, W.; Schwarz, H.; Peschel, A.; Götz, F. Role of staphylococcal wall teichoic acid in targeting the major autolysin Atl. Mol. Microbiol. 2010, 75, 864–873. [Google Scholar] [CrossRef]
- Covas, G.; Vaz, F.; Henriques, G.; Pinho, M.G.; Filipe, S.R. Analysis of Cell Wall Teichoic Acids in Staphylococcus aureus. Methods Mol. Biol. 2016, 1440, 201–213. [Google Scholar] [CrossRef]
- Rouser, G.; Fleischer, S.; Yamamoto, A. Two dimensional thin layer chromatographic separation of polar lipids and determination of phospholipids by phosphorus analysis of spots. Lipids 1970, 5, 494–496. [Google Scholar] [CrossRef]
- Kühner, D.; Stahl, M.; Demircioglu, D.D.; Bertsche, U. From cells to muropeptide structures in 24 h: Peptidoglycan mapping by UPLC-MS. Sci. Rep. 2014, 4, 7494. [Google Scholar] [CrossRef] [Green Version]
- de Jonge, B.L.; Chang, Y.S.; Gage, D.; Tomasz, A. Peptidoglycan composition of a highly methicillin-resistant Staphylococcus aureus strain. The role of penicillin binding protein 2A. J. Biol. Chem. 1992, 267, 11248–11254. [Google Scholar] [PubMed]
- Gajdiss, M.; Türck, M.; Bierbaum, G. Bacterial Histidine Kinases: Overexpression, Purification, and Inhibitor Screen. Methods Mol. Biol. 2017, 1520, 247–259. [Google Scholar] [CrossRef] [PubMed]
- Türck, M.; Bierbaum, G. Purification and activity testing of the full-length YycFGHI proteins of Staphylococcus aureus. PLoS ONE 2012, 7, e30403. [Google Scholar] [CrossRef] [PubMed]
- Sabala, I.; Jonsson, I.-M.; Tarkowski, A.; Bochtler, M. Anti-staphylococcal activities of lysostaphin and LytM catalytic domain. BMC Microbiol. 2012, 12, 97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pestka, S.; Daugherty, B.L.; Jung, V.; Hotta, K.; Pestka, R.K. Anti-mRNA: Specific inhibition of translation of single mRNA molecules. Proc. Natl. Acad. Sci. USA 1984, 81, 7525–7528. [Google Scholar] [CrossRef] [Green Version]
- Delaune, A.; Poupel, O.; Mallet, A.; Coic, Y.-M.; Msadek, T.; Dubrac, S. Peptidoglycan crosslinking relaxation plays an important role in Staphylococcus aureus WalKR-dependent cell viability. PLoS ONE 2011, 6, e17054. [Google Scholar] [CrossRef] [Green Version]
- Hardt, P.; Engels, I.; Rausch, M.; Gajdiss, M.; Ulm, H.; Sass, P.; Ohlsen, K.; Sahl, H.-G.; Bierbaum, G.; Schneider, T.; et al. The cell wall precursor lipid II acts as a molecular signal for the Ser/Thr kinase PknB of Staphylococcus aureus. Int. J. Med. Microbiol. 2017, 307, 1–10. [Google Scholar] [CrossRef]
- Libby, E.A.; Goss, L.A.; Dworkin, J. The Eukaryotic-Like Ser/Thr Kinase PrkC Regulates the Essential WalRK Two-Component System in Bacillus subtilis. PLoS Genet. 2015, 11, e1005275. [Google Scholar] [CrossRef]
- Poupel, O.; Proux, C.; Jagla, B.; Msadek, T.; Dubrac, S. SpdC, a novel virulence factor, controls histidine kinase activity in Staphylococcus aureus. PLoS Pathog. 2018, 14, e1006917. [Google Scholar] [CrossRef] [Green Version]
- Howell, A.; Dubrac, S.; Andersen, K.K.; Noone, D.; Fert, J.; Msadek, T.; Devine, K. Genes controlled by the essential YycG/YycF two-component system of Bacillus subtilis revealed through a novel hybrid regulator approach. Mol. Microbiol. 2003, 49, 1639–1655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Botella, E.; Devine, S.K.; Hubner, S.; Salzberg, L.I.; Gale, R.T.; Brown, E.D.; Link, H.; Sauer, U.; Codée, J.D.; Noone, D.; et al. PhoR autokinase activity is controlled by an intermediate in wall teichoic acid metabolism that is sensed by the intracellular PAS domain during the PhoPR-mediated phosphate limitation response of Bacillus subtilis. Mol. Microbiol. 2014, 94, 1242–1259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fritz, G.; Mascher, T. A balancing act times two: Sensing and regulating cell envelope homeostasis in Bacillus subtilis. Mol. Microbiol. 2014, 94, 1201–1207. [Google Scholar] [CrossRef] [PubMed]
- Atilano, M.L.; Pereira, P.M.; Vaz, F.; Catalão, M.J.; Reed, P.; Grilo, I.R.; Sobral, R.G.; Ligoxygakis, P.; Pinho, M.G.; Filipe, S.R. Bacterial autolysins trim cell surface peptidoglycan to prevent detection by the Drosophila innate immune system. Elife 2014, 3, e02277. [Google Scholar] [CrossRef]
- Dubrac, S.; Bisicchia, P.; Devine, K.M.; Msadek, T. A matter of life and death: Cell wall homeostasis and the WalKR (YycGF) essential signal transduction pathway. Mol. Microbiol. 2008, 70, 1307–1322. [Google Scholar] [CrossRef]
- Rausch, M.; Deisinger, J.P.; Ulm, H.; Müller, A.; Li, W.; Hardt, P.; Wang, X.; Li, X.; Sylvester, M.; Engeser, M.; et al. Coordination of capsule assembly and cell wall biosynthesis in Staphylococcus aureus. Nat. Commun. 2019, 10, 1404. [Google Scholar] [CrossRef]
- Kim, T.; Choi, J.; Lee, S.; Yeo, K.J.; Cheong, H.-K.; Kim, K.K. Structural Studies on the Extracellular Domain of Sensor Histidine Kinase YycG from Staphylococcus aureus and Its Functional Implications. J. Mol. Biol. 2016, 428, 3074–3089. [Google Scholar] [CrossRef]
- Wu, R.; Gu, M.; Wilton, R.; Babnigg, G.; Kim, Y.; Pokkuluri, P.R.; Szurmant, H.; Joachimiak, A.; Schiffer, M. Insight into the sporulation phosphorelay: Crystal structure of the sensor domain of Bacillus subtilis histidine kinase, KinD. Protein Sci. 2013, 22, 564–576. [Google Scholar] [CrossRef] [Green Version]
- Reinelt, S.; Hofmann, E.; Gerharz, T.; Bott, M.; Madden, D.R. The structure of the periplasmic ligand-binding domain of the sensor kinase CitA reveals the first extracellular PAS domain. J. Biol. Chem. 2003, 278, 39189–39196. [Google Scholar] [CrossRef] [Green Version]
- Cheung, J.; Hendrickson, W.A. Crystal structures of C4-dicarboxylate ligand complexes with sensor domains of histidine kinases DcuS and DctB. J. Biol. Chem. 2008, 283, 30256–30265. [Google Scholar] [CrossRef] [Green Version]
- Türck, M. Das essentielle YycFGHI-Regulationssystem von Staphylococcus aureus: Charakterisierung von Überexpressions-Mutanten und die Etablierung zweier in vitro-Modellsysteme. Ph.D. Thesis, University of Bonn, Bonn, Germany, July 2009. [Google Scholar]
- Büttner, F.M.; Zoll, S.; Nega, M.; Götz, F.; Stehle, T. Structure-function analysis of Staphylococcus aureus amidase reveals the determinants of peptidoglycan recognition and cleavage. J. Biol. Chem. 2014, 289, 11083–11094. [Google Scholar] [CrossRef] [Green Version]
- Gammoh, N.Z.; Rink, L. Zinc in Infection and Inflammation. Nutrients 2017, 9, 624. [Google Scholar] [CrossRef] [PubMed] [Green Version]




| Primer | 5′-3′ sequence 2 | Restriction Site |
|---|---|---|
| AS-yycH_for | GAATCTAGAACGCATGTTTTTGCACCA | XbaI |
| AS-yycH_rev | ATGGAATTCTCGCTTCATCTTCGGACA | EcoRI |
| AS-yycI_for | GTTCTAGAGCGCGTATTTAAAGGTGCT | XbaI |
| AS-yycI_rev | TATGAATTCGCACCATCTGTGGGCTTA | EcoRI |
| AS-yycHI_for | CAGTCTAGACGTACCGCGTTGGTATGT | XbaI |
| AS-yycHI_rev | CTTGAATTCTGTGTGAGCGATTGACTTT | EcoRI |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gajdiss, M.; Monk, I.R.; Bertsche, U.; Kienemund, J.; Funk, T.; Dietrich, A.; Hort, M.; Sib, E.; Stinear, T.P.; Bierbaum, G. YycH and YycI Regulate Expression of Staphylococcus aureus Autolysins by Activation of WalRK Phosphorylation. Microorganisms 2020, 8, 870. https://doi.org/10.3390/microorganisms8060870
Gajdiss M, Monk IR, Bertsche U, Kienemund J, Funk T, Dietrich A, Hort M, Sib E, Stinear TP, Bierbaum G. YycH and YycI Regulate Expression of Staphylococcus aureus Autolysins by Activation of WalRK Phosphorylation. Microorganisms. 2020; 8(6):870. https://doi.org/10.3390/microorganisms8060870
Chicago/Turabian StyleGajdiss, Mike, Ian R. Monk, Ute Bertsche, Janina Kienemund, Tanja Funk, Alina Dietrich, Michael Hort, Esther Sib, Timothy P. Stinear, and Gabriele Bierbaum. 2020. "YycH and YycI Regulate Expression of Staphylococcus aureus Autolysins by Activation of WalRK Phosphorylation" Microorganisms 8, no. 6: 870. https://doi.org/10.3390/microorganisms8060870