X-ray Crystal Structure, Geometric Isomerism, and Antimicrobial Activity of New Copper(II) Carboxylate Complexes with Imidazole Derivatives
Abstract
:1. Introduction
2. Results and Discussion
2.1. Synthesis of the Complexes
2.2. Characterization of the Complexes
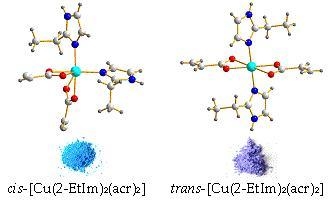
2.2.1. Description of the X-ray Crystal Structures of the Complexes
2.2.2. Infrared Spectra
2.2.3. UV-Vis-NIR Spectral Data
2.2.4. Electron Paramagnetic Resonance Spectral Data
2.2.5. Morphology Studies by Scanning Electron Microscopy
2.2.6. Mass Spectrometry
2.2.7. Powder X-ray Diffraction
2.2.8. Antimicrobial Assay
3. Experimental Section
3.1. General Information
3.2. Synthesis of Complexes
3.3. Biological Assays
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Melník, M.; Auderová, M.; Hol'ko, M. Copper(II) carboxylates and their antimicrobial effect. Inorg. Chim Acta 1982, 67, 117–120. [Google Scholar] [CrossRef]
- Mojumdar, S.C.; Madgurambal, G.; Saleh, M.T. A study on synthesis and thermal, spectral and biological properties of carboxylato-Mg(II) and carboxylato-Cu(II) complexes with bioactive ligands. J. Therm. Anal. Calorim. 2005, 81, 205–210. [Google Scholar] [CrossRef]
- Kozlevčar, B.; Leban, I.; Turel, I.; Šegedin, P.; Petric, M.; Pohleven, F.; White, A.; Williams, D.; Sieler, J. Complexes of copper(II) acetate with nicotinamide: Preparation, characterization and fungicidal activity; crystal structures of [Cu2(O2CCH3)4(nia)] and [Cu2(O2CCH3)4(nia)2]. Polyhedron 1999, 18, 755–762. [Google Scholar] [CrossRef]
- Ranford, J.D.; Sadler, P.J.; Tocher, D.A. Cytotoxicity and antiviral activity of transition-metal salicylato complexes and crystal structure of Bis(diisopropylsalicylato)(1,10-phenanthroline)copper(II). J. Chem. Soc. Dalton Trans. 1993, 22, 3393–3399. [Google Scholar] [CrossRef]
- Wang, Q.; Yu, Z.; Wang, Q.; Li, W.; Gao, F.; Li, S. Synthesis, crystal structure and DNA-binding properties of a mononuclear copper complex with pyridine-2-carboxylate ligand. Inorg. Chim. Acta 2012, 383, 230–234. [Google Scholar] [CrossRef]
- O’Connor, M.; Kellett, A.; McCann, M.; Rosair, G.; McNamara, M.; Howe, O.; Creaven, B.S.; McClean, S.; Foltyn-Arfa Kia, A.; O’Shea, D.; et al. Copper(II) complexes of salicylic acid combining superoxide dismutase mimetic properties with DNA binding and cleaving capabilities display promising chemotherapeutic potential with fast acting in vitro cytotoxicity against cisplatin sensitive and resistant cancer cell lines. J. Med. Chem. 2012, 55, 1957–1968. [Google Scholar] [PubMed]
- Devereux, M.; O’Shea, D.; O’Connor, M.; Grehan, H.; Connor, G.; McCann, M.; Rosair, G.; Lyng, F.; Kellett, A.; Walsh, M.; et al. Synthesis, catalase, superoxide dismutase and antitumour activities of copper(II) carboxylate complexes incorporating benzimidazole, 1,10-phenanthroline and bipyridine ligands: X-ray crystal structures of [Cu(BZA)2(bipy)(H2O)], [Cu(SalH)2(BZDH)2] and [Cu(CH3COO)2(5,6-DMBZDH)2] (SalH2 = salicylic acid; BZAH = benzoic acid; BZDH = benzimidazole and 5,6-DMBZDH = 5,6-dimethylbenzimidazole). Polyhedron 2007, 26, 4073–4084. [Google Scholar]
- Kreuder, J.; Otten, A.; Fuder, H.; Tønnesen, T.; Horn, N.; Dralle, D. Clinical and biochemical consequences of copper-histidine therapy in Menkes disease. Eur. J. Pediatr. 1993, 152, 828–832. [Google Scholar] [CrossRef]
- Zhou, L.J.; Luan, X.J.; Wang, Y.Y.; Lee, G.H.; Shi, Q.Z.; Peng, S.M. Supramolecular complexes constructed with carboxylate Cu(II) and 2-(2-pyridyl)-benzimidazole via hydrogen bonding. J. Coord. Chem. 2006, 59, 1107–1121. [Google Scholar] [CrossRef]
- Ye, B.H.; Tong, M.L.; Chen, X.M. Metal-organic molecular architectures with 2,2′-bipyridyl-like and carboxylate ligands. Coord. Chem. Rev. 2005, 249, 545–565. [Google Scholar] [CrossRef]
- Gomathi, S.; Muthiah, P.T. Supramolecular architecture of metal-organic frameworks involving dinuclear copper paddle-wheel complexes. Acta Crystallogr. C. 2013, 69, 1498–1502. [Google Scholar] [CrossRef] [PubMed]
- Moulton, B.; Zaworotko, M. From molecules to crystal engineering: supramolecular isomerism and polymorphism in network solids. Chem. Rev. 2001, 101, 1629–1658. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.; Pandurangan, A.; Rana, K.; Anand, P.; Ahamad, A.; Tiwari, A.K. Benzimidazole: A short review of their antimicrobial activities. Int. Curr. Pharm. J. 2011, 5, 119–127. [Google Scholar] [CrossRef]
- Ülküseven, B.; Tavman, A.; Ötük, G. Synthesis, characterization and antimicrobial activity of d8-10 metal complexes of some 2-Substituted-1H-Benzimidazoles. Met. Based Drugs 1999, 6, 163–167. [Google Scholar] [CrossRef] [PubMed]
- Sigel, H. Ternary complexes in solution. 34. Discriminating and stability increasing properties of the imidazole moiety in mixed-ligand complexes. Inorg. Chem. 1980, 19, 1411–1413. [Google Scholar] [CrossRef]
- Bernarducci, E.E.; Bharadwaj, P.K.; Lalancette, R.A.; Korgh-Jesperson, K.; Potenza, J.A.; Schugar, M.J. Molecular structures, electronic spectra, and ESR spectra of bis(4,4’,5,5’-tetramethyl-2,2’-biimidazole)copper(II) dinitrate and bis(4,4’,5,5’-tetramethyl-2,2’-biimidazole)zinc(II)0.90copper(II)0.10 dinitrate. Inorg. Chem. 1983, 22, 3911–3920. [Google Scholar] [CrossRef]
- Wang, Y.Y.; Shi, Q.; Shi, Q.Z.; Gao, Y.C.; Zhou, Z.Y. Syntheses, characterization and crystal structure of copper(II) α,β-unsaturated carboxylate complexes with imidazole. Polyhedron 1999, 18, 2009–2015. [Google Scholar] [CrossRef]
- Wang, Y.Y.; Zhou, L.J.; Shi, Q.; Shi, Q.Z.; Gao, Y.C.; Hou, X. Novel trinuclear copper(II) complexes with α,β-unsaturated carboxylates and imidazole. Trans. Met. Chem. 2002, 27, 145–148. [Google Scholar] [CrossRef]
- Jian, F.; Wang, Z.; Bai, Z.; You, X.; Chen, W. Synthesis, structural and spectroscopic characterization of the α and β forms of bis(imidazole)copper(II) dibenzoate, [Cu(im)2(OBz)2]. Trans. Met. Chem. 1999, 24, 589–594. [Google Scholar] [CrossRef]
- Abuhijleh, A.L.; Woods, C. Syntheses and molecular structures of cis and trans monomeric ferrocenecarboxylatocopper(II) complexes with imidazole derivatives. J. Chem. Soc. Dalton Trans. 1992, 7, 1249–1252. [Google Scholar] [CrossRef]
- Vlaicu, I.D.; Constand, M.; Olar, R.; Marinescu, D.; Grecu, M.N.; Lazar, V.; Chifiriuc, M.C.; Badea, M. Thermal stability of new biologic active copper(II) complexes with 5,6-dimethylbenzimidazole. J. Therm. Anal. Calorim. 2013, 113, 1369–1377. [Google Scholar] [CrossRef]
- Vlaicu, I.D.; Olar, R.; Marinescu, D.; Lazar, V.; Badea, M. Physico-chemical and thermal characterisation of new Co(II) complexes with pyrazole derivatives. J. Therm. Anal. Calorim. 2013, 113, 1337–1343. [Google Scholar] [CrossRef]
- Badea, M.; Vlaicu, I.D.; Olar, R.; Constand, M.; Bleotu, C.; Chifiriuc, M.C.; Marutescu, L.; Lazar, V.; Grecu, M.N.; Marinescu, D. Thermal behaviour and characterisation of new biologically active Cu(II) complexes with benzimidazole as main ligand. J. Therm. Anal. Calorim. 2014, 118, 1119–1126. [Google Scholar] [CrossRef]
- Mrozinski, J.; Heyduk, E. Binuclear copper(II) complexes with α,β-unsaturated carboxylic acids. 1. Copper acrylates with heterocyclic amine adducts. Polish J. Chem. 1982, 56, 683–689. [Google Scholar]
- Tong, Y.P.; Zheng, S.L. Synthesis, structure, spectroscopic properties, DFT and TDDFT investigations of copper(II) complex with 2-(2-hydroxyphenyl)benzimidazole. J. Mol. Struct. 2007, 841, 34–40. [Google Scholar] [CrossRef]
- Saczewski, F.; Dziemidowicz-Borys, E.; Bednarski, P.J.; Grunert, R.; Gdaniec, M.; Tabin, P. Synthesis, crystal structure and biological activities of copper(II) complexes with chelating bidentate 2-substituted benzimidazole ligands. J. Inorg. Biochem. 2006, 100, 1389–1398. [Google Scholar] [CrossRef] [PubMed]
- Hathaway, B.J. A new look at the stereochemistry and electronic properties of complexes of the copper(II) ion. In Complex Chemistry (Structure and Bonding); Emsley, J., Ed.; Springer: Berlin/Heidelberg, Germany, 1984; Volume 57, pp. 55–118. [Google Scholar]
- Hathaway, B.J.; Tomlinson, A.A.G. Copper(II) ammonia complexes. Coord. Chem. Rev. 1970, 5, 1–43. [Google Scholar] [CrossRef]
- Hathaway, B.J.; Billing, D.E. The electronic properties and stereochemistry of mono-nuclear complexes of the copper(II) ion. Coord. Chem. Rev. 1970, 5, 143–207. [Google Scholar] [CrossRef]
- Morzyk-Ociepa, B.; Różycka-Sokołowska, E.; Michalska, D. Revised crystal and molecular structure, FT-IR spectra and DFT studies of chlorotetrakis(imidazole)copper(II) chloride. J. Mol. Struct. 2012, 1028, 49–56. [Google Scholar] [CrossRef]
- Deacon, G.B.; Philips, J.R. Relationships between the carbon-oxygen stretching frequencies of carboxylato complexes and the type of carboxylate coordination. Coord. Chem. Rev. 1980, 33, 227–250. [Google Scholar] [CrossRef]
- Oldham, C. Carboxylates, squarates and related species. In Comprehensive Coordination Chemistry; Wilkinson, G., Gillard, R.D., McCleverty, J.A., Eds.; Pergamon Press: Oxford, UK, 1987; Volume 2, pp. 435–460. [Google Scholar]
- Lever, A.B.P. Inorganic Electronic Spectroscopy, 2nd ed.; Elsevier: New York, NY, USA, 1984; pp. 553–572. [Google Scholar]
- Smith, D.W. Relationship between electron spin resonance g-values and covalent bonding in tetragonal copper(II) compounds. J. Chem. Soc. A 1970, 3108–3120. [Google Scholar] [CrossRef]
- Henderson, W.; McIndoe, J.S. Mass Spectrometry of Inorganic, Coordination and Organometallic Compounds; John Wiley & Sons: West Sussex, UK, 2005; pp. 127–167. [Google Scholar]
- Coculescu, B.-I.; Palade, A.-M.; Delcaru, C.; Coculescu, E.C. Genetic analysis of multidrug-resistant salmonella enterica serovar typhimurium strains producing extended-spectrum Β-lactamases (ESBL) associated with diarrhea in romanian pediatric patients. Rom. Biotechnol. Lett. 2016, 21, 11393–11403. [Google Scholar]
- Merezeanu, N.; Gheorghe, I.; Popa, M.; Lazăr, V.; Banu, O.; Bolocan, A.; Grigore, R.; Vifor Berteșteanu, Ș.; Pântea, O. Phenotypic and genotypic investigation of resistance and virulence features of methicillin resistant Staphylococcus aureus strains isolated from hospitalized patients. Rom. Biotechnol. Lett. 2016, 21, 11591–11598. [Google Scholar]
- Georgescu, M.; Vrinceanu, D.; Radulescu, L.; Tusaliu, M.; Martu, C.; Curutiu, C.; Hussien, M.D.; Budu, V. Microbial biofilms and implantable hearing aids. Rom. Biotechnol. Lett. 2017, 22, 12681–12686. [Google Scholar]
- De los Ríos, A.; Pérez-Ortega, S.; Wierzchos, J.; Ascaso, C. Differential effects of biocide treatments on saxicolous communities: Case study of the Segovia cathedral cloister (Spain). Int. Biodeterior. Biodegrad. 2012, 67, 64–72. [Google Scholar] [CrossRef] [Green Version]
- Scheerer, S.; Ortega-Morales, O.; Gaylarde, C. Microbial deterioration of stone monuments-an updated overview. Adv. Appl. Microbiol. 2009, 66, 97–139. [Google Scholar] [PubMed]
- Olar, R.; Badea, M.; Grecu, M.; Marinescu, D.; Lazar, V.; Balotescu, C. Copper(II) complexes with N,N-dimethylbiguanide. J. Therm. Anal. Calorim. 2008, 92, 239–243. [Google Scholar] [CrossRef]
- Pahonțu, E.; Ilieș, D.-C.; Shova, S.; Paraschivescu, C.; Badea, M.; Gulea, A.; Roșu, T. Synthesis, characterization, crystal structure and antimicrobial activity of copper(II) complexes with the schiff base derived from 2-Hydroxy-4-Methoxybenzaldehyde. Molecules 2015, 20, 5771–5792. [Google Scholar] [CrossRef]
- Liu, H.; Yang, W.; Zhou, W.; Xu, Y.; Xie, J.; Li, M. Crystal structures and antimicrobial activities of copper(II) complexes of fluorine-containing thioureido ligands. Inorg. Chim. Acta 2013, 405, 387–394. [Google Scholar] [CrossRef]
- Gupta, S.P.; Sharma, K. The role of fungi in biodeterioration of sandstone with reference to Mahadev temple, Bastar, Chhatisgarh. Recent Res. Sci. Technol. 2012, 4, 18–21. [Google Scholar]
- Sirghi, A.; Gheorghe, I.; Marutescu, L.; Batalu, D.; Badica, P.; Badea, M.; Olar, R.; Sadik, O.; Aziz, G.A.; Avram, I.; et al. Activity of different inorganic nanoparticles against fungal isolates colonising buildings included in the Romanian National Heritage. Rev. Română Med. Lab. 2018, 26, S40. [Google Scholar]
- Dolomanov, O.V.; Bourhis, L.J.; Gildea, R.J.; Howard, J.A.K.; Puschmann, H. OLEX2: A complete structure solution, refinement and analysis program. J. Appl. Cryst. 2009, 42, 339–341. [Google Scholar] [CrossRef]
- Sheldrick, G.M. A short history of SHELX. Acta Crystallogr. A. 2008, 64, 112–122. [Google Scholar] [CrossRef]
- Sheldrick, G.M. Crystal structure refinement with SHELXL. Acta Crystallogr. C Struct. Chem. 2015, 71, 3–8. [Google Scholar] [CrossRef]
- APEX3, Bruker AXS Inc.: Madison, WI, USA, 2016.
- XP–Interactive molecular graphics, Version 5.1; Bruker AXS Inc.: Madison, WI, USA, 1998.
- Stoll, S.; Schweiger, A. EasySpin, a comprehensive software package for spectral simulation and analysis in EPR. J. Magn. Reson. 2006, 178, 42–55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Sample Availability: Samples of all compounds are available from the authors. |








| Compound | [Cu(2-MeIm)2(acr)2] (2) | [Cu(2-EtIm)2(acr)2] (4) | [Cu(2-EtIm)2(acr)2] (5) |
|---|---|---|---|
| Empirical formula | C14H18CuN4O4 | C16H22CuN4O4 | C16H22CuN4O4 |
| Formula weight | 369.86 | 397.92 | 397.92 |
| Temperature/K | 296(2) | 296(2) | 296(2) |
| Crystal system | monoclinic | monoclinic | monoclinic |
| Space group | P21/c | C2/c | P21/c |
| a/Å | 7.6597(3) | 12.9665(7) | 7.79135(13) |
| b/Å | 7.6111(2) | 9.5520(4) | 7.757740(13) |
| c/Å | 15.0825(6) | 16.6741(9) | 15.6222(3) |
| α/° | 90.00 | 90.00 | 90.00 |
| β/° | 104.266(4) | 114.025(7) | 104.2293(18) |
| γ/° | 90.00 | 90.00 | 90.00 |
| Volume /Å | 852.2(1) | 1886.3(2) | 915.3(1) |
| Z | 2 | 4 | 2 |
| Calculated density /mg/mm3 | 1.441 | 1.401 | 1.444 |
| Absorption coefficient /mm–1 | 2.030 | 1.873 | 1.930 |
| F(000) | 382 | 828 | 414 |
| No. of measured and independent reflections | 2664/1605 | 2969/1792 | 9556/1779 |
| Rint | 0.010 | 0.027 | 0.029 |
| No. of parameters/restraints | 111/1 | 119/1 | 148/66 |
| Goodness-of-fit on F2 | 1.105 | 1.085 | 1.056 |
| Final R indexes (I ≥ 2σ (I)) | R1 = 0.029 wR2 = 0.085 | R1 = 0.049 wR2 = 0.138 | R1 = 0.029 wR2 = 0.084 |
| Final R indexes (all data) | R1 = 0.032 wR2 = 0.087 | R1 = 0.054 wR2 = 0.148 | R1 = 0.031 wR2 = 0.086 |
| Largest diff. peak/hole/e Å−3 | 0.23/−0.27 | 0.48/−0.97 | 0.19/−0.39 |
| CCDC Nr. | 1841956 | 1841958 | 1841957 |
| Bonds | (2) | (4) | (5) |
|---|---|---|---|
| Cu1–O1 | 1.969(1) | 1.982(2) | 1.960(1) |
| Cu1–O1* | 1.969(1) | 1.982(2) | 1.960(1) |
| Cu1–O2 | 2.672(1) | 2.605(2) | 2.857(9) |
| Cu1–O2* | 2.672(1) | 2.605(2) | 2.857(9) |
| Cu1–N1 | 1.986(1) | 1.985(2) | 1.989(1) |
| Cu1–N1* | 1.986(1) | 1.985(2) | 1.989(1) |
| O1–Cu1–N1 | 91.1(1) | 90.3(1) | 89.0(1) |
| O1–Cu1–N1* | 89.0(1) | 170.9(1) | 91.0(1) |
| O1*–Cu1–N1* | 91.1(1) | 90.3(1) | 89.0(1) |
| N1–Cu1–N1* | 180 | 93.5(1) | 180 |
| O1–Cu1–O1* | 180 | 87.1(1) | 180 |
| O2–Cu1–O2* | 180 | 127.9(1) | 180 |
| O2–Cu1–N1 | 90.8(1) | 99.8(1) | 89.7(3) |
| O2–Cu1–N1* | 89.2(1) | 115.5(1) | 90.3(3) |
| O2–Cu1–O1* | 125.8(1) | 85.9(1) | 128.0(2) |
| O1–Cu1–O2* | 125.8(1) | 85.9(1) | 128.0(2) |
| O1–Cu1–O2 | 54.2(1) | 55.6(1) | 52.0(2) |
| O1*–Cu1–O2* | 54.2(1) | 55.6(1) | 52.0(2) |
| O2*–Cu1–N1* | 90.8(1) | 99.8(1) | 89.7(3) |
| N1–Cu1–O1* | 89.0(1) | 170.9(1) | 91.0(1) |
| N1–Cu1–O2* | 89.2(1) | 115.5(1) | 90.3(2) |
| Symmetry operations | (*) 1−x, 1−y, 1−z | (*) 2−x, y, 1.5−z | (*) −x, −y, 1−z |
| Complex | D–H (Å) N2–H2A | H…A (Å) H2A…O2 | D…A (Å) N2…O2 | <(DHA) (°) <(N2–H2A…O2) |
|---|---|---|---|---|
| (2) (a) | 0.88(2) | 1.93(2) | 2.784(2) | 165(3) |
| (4) (b) | 0.87(2) | 1.95(2) | 2.801(3) | 166(4) |
| (5) (c) | 0.85(2) | 1.99(2) | 2.797(1) | 171(3) |
| 2-MeIm | 5-MeIm | 2-EtIm | (1) | (2) | (3) | (4) | (5) | Assignments |
|---|---|---|---|---|---|---|---|---|
| - | - | - | 3405 s | - | - | - | - | ν(OH2) |
| 3136 m | 3136 m | 3153 w | 3116 s | 3125 m | 3131 w | 3140 m | 3134 m | ν(CH), ν(NH) |
| - | - | - | 2944 w | 2915 w | 2925 w | 2920 w | 2925 w | νas(CH2) |
| - | - | - | 2880 w | 2885 w | 2888 w | 2880 w | 2885 w | νs(CH2) |
| 1676 m | 1676 m | 1673 w | 1676 w | 1630 m | 1625 m | 1658 w | - | ν(C=N) |
| 1596 vs | 1596 vs | - | 1613 vs | 1605 vs | - | 1615 s | 1609 vs | δ(NH), ν(CC), ν(CN) |
| - | - | - | 1551 s | 1568 vs | 1576 vs | 1565 vs | 1587 vs | νas(COO) |
| - | - | - | 1503 s | 1515 m | 1520 m | 1525 m | 1542 m | ν(C=C) aliph |
| - | - | - | 1368 s | 1416 m | 1387 s | 1380 m | 1410 vs | νs(COO) |
| - | - | - | 1283 s | 1280 w | 1290 w | 1280 m | 1285 m | δ(CH2) |
| 1206 w | 1206 w | 1243 w | 1228 s | 1215 w | 1200 w | 1240 w | 1245 w | ν(CN), δ(CH) |
| 1155 vs | 1155 vs | 1153 m | 1153 m | 1135 w | 1112 vs | 1158 m | 1163 m | ν(CC), ν(CN), δ(CH) |
| 942 s | 942 s | 956 s | 956 m | 955 m | 963 m | 950 w | 955 w | δ(CH), δ(imidazole ring) |
| 875 w | 875 w | 875 w | 864 m | 870 m | 875 m | 870 m | 879 m | π(CH), δ(imidazole ring) |
| 756 vs | 756 vs | 751 vs | 779 s | 760 s | - | 755 m | 758 m | π(CH) |
| 627 w | 627 w | 625 w | 647 m | 650 m | 625 m | 630 m | 624 m | π(NH) |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vlaicu, I.D.; Borodi, G.; Scăețeanu, G.V.; Chifiriuc, M.C.; Măruțescu, L.; Popa, M.; Stefan, M.; Mercioniu, I.F.; Maurer, M.; Daniliuc, C.G.; et al. X-ray Crystal Structure, Geometric Isomerism, and Antimicrobial Activity of New Copper(II) Carboxylate Complexes with Imidazole Derivatives. Molecules 2018, 23, 3253. https://doi.org/10.3390/molecules23123253
Vlaicu ID, Borodi G, Scăețeanu GV, Chifiriuc MC, Măruțescu L, Popa M, Stefan M, Mercioniu IF, Maurer M, Daniliuc CG, et al. X-ray Crystal Structure, Geometric Isomerism, and Antimicrobial Activity of New Copper(II) Carboxylate Complexes with Imidazole Derivatives. Molecules. 2018; 23(12):3253. https://doi.org/10.3390/molecules23123253
Chicago/Turabian StyleVlaicu, Ioana Dorina, Gheorghe Borodi, Gina Vasile Scăețeanu, Mariana Carmen Chifiriuc, Luminița Măruțescu, Marcela Popa, Mariana Stefan, Ionel Florinel Mercioniu, Martin Maurer, Constantin G. Daniliuc, and et al. 2018. "X-ray Crystal Structure, Geometric Isomerism, and Antimicrobial Activity of New Copper(II) Carboxylate Complexes with Imidazole Derivatives" Molecules 23, no. 12: 3253. https://doi.org/10.3390/molecules23123253