Molecular Features and Metal Ions That Influence 10-23 DNAzyme Activity
Abstract
:1. Introduction
1.1. Biological Aspects and Therapeutic Potential of the 10-23 DNAzyme
1.2. Structural Information on DNAzymes
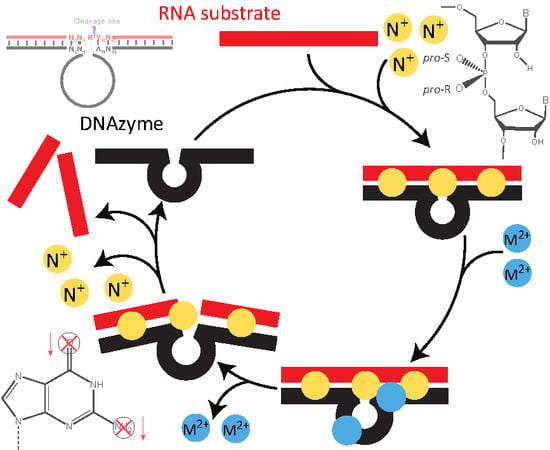
1.3. Kinetics of DNAzyme-Mediated Catalysis
2. Impact of Nucleotide Substitutions within the Binding Arms on Catalytic Activity
DNAzyme Activity Depends Upon Nucleotides Flanking the Cleavage Site
3. Impact of Deoxynucleotide Substitutions and Modifications within the Catalytic Loop on Activity
3.1. The Deoxynucleotides at Position 1 to 6, 13, and 14 Are Crucial for Activity
3.2. Exocyclic Functional Groups at Position 6 and 14 Are Crucial for the Activity of the 10-23 DNAzyme
3.3. The Exocyclic Amino Groups at dC and dC Display Additional Functional Importance
3.4. Non-Bridging Oxygen Atoms of the Phosphate Group between dT and dA May Be Involved in Metal Ion Coordination
3.5. Deletion of the Deoxynucleotide dT Leads to an Active 10-23 DNAzyme
3.6. Adenine Minor Groove Interactions Play a Role in 10-23 DNAzyme-Mediated Catalysis
3.7. A DNAzyme Variant with an 11 Nt-Containing Catalytic Loop Requires Ca for Its Activity
4. Influence of Metal Ions on 10-23 DNAzyme Activity
5. Critical DNAzyme Positions and Their Potential Involvement in DNAzyme Catalysis
6. Conflicting Results
7. Conclusions
Funding
Conflicts of Interest
References
- Santoro, S.W.; Joyce, G.F. A general purpose RNA-cleaving DNA enzyme. Proc. Natl. Acad. Sci. USA 1997, 94, 4262–4266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flynn-Charlebois, A.; Wang, Y.; Prior, T.K.; Rashid, I.; Hoadley, K.A.; Coppins, R.L.; Wolf, A.C.; Silverman, S.K. Deoxyribozymes with 2′-5′ RNA ligase activity. J. Am. Chem. Soc. 2003, 125, 2444–2454. [Google Scholar] [CrossRef] [PubMed]
- Chandra, M.; Sachdeva, A.; Silverman, S.K. DNA-catalyzed sequence-specific hydrolysis of DNA. Nat. Chem. Biol. 2009, 5, 718–720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cuenoud, B.; Szostak, J.W. A DNA metalloenzyme with DNA ligase activity. Nature 1995, 375, 611–614. [Google Scholar] [CrossRef]
- Chinnapen, D.J.F.; Sen, D. A deoxyribozyme that harnesses light to repair thymine dimers in DNA. Proc. Natl. Acad. Sci. USA 2004, 101, 65–69. [Google Scholar] [CrossRef] [Green Version]
- Silverman, S.K. Pursuing DNA catalysts for protein modification. Acc. Chem. Res. 2015, 48, 1369–1379. [Google Scholar] [CrossRef] [Green Version]
- Wong, O.Y.; Pradeepkumar, P.; Silverman, S.K. DNA-catalyzed covalent modification of amino acid side chains in tethered and free peptide substrates. Biochemistry 2011, 50, 4741–4749. [Google Scholar] [CrossRef] [Green Version]
- Pradeepkumar, P.; Höbartner, C.; Baum, D.A.; Silverman, S.K. DNA-catalyzed formation of nucleopeptide linkages. Angew. Chem. Int. Ed. 2008, 47, 1753–1757. [Google Scholar] [CrossRef]
- Chu, C.C.; Wong, O.Y.; Silverman, S.K. A generalizable DNA-catalyzed approach to peptide–nucleic acid conjugation. ChemBioChem 2014, 15, 1905–1910. [Google Scholar] [CrossRef] [Green Version]
- Chandrasekar, J.; Silverman, S.K. Catalytic DNA with phosphatase activity. Proc. Natl. Acad. Sci. USA 2013, 110, 5315–5320. [Google Scholar] [CrossRef] [Green Version]
- Victor, J.; Steger, G.; Riesner, D. Inability of DNAzymes to cleave RNA in vivo is due to limited Mg2+ concentration in cells. Eur. Biophys. J. 2018, 47, 333–343. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Dass, C.R.; Sumithran, E.; Di Girolamo, N.; Sun, L.Q.; Khachigian, L.M. Effect of deoxyribozymes targeting c-Jun on solid tumor growth and angiogenesis in rodents. J. Natl. Cancer Inst. 2004, 96, 683–696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Gasper, W.J.; Stass, S.A.; Ioffe, O.B.; Davis, M.A.; Mixson, A.J. Angiogenic inhibition mediated by a DNAzyme that targets vascular endothelial growth factor receptor 2. Cancer Res. 2002, 62, 5463–5469. [Google Scholar] [PubMed]
- Sel, S.; Wegmann, M.; Dicke, T.; Sel, S.; Henke, W.; Yildirim, A.Ö.; Renz, H.; Garn, H. Effective prevention and therapy of experimental allergic asthma using a GATA-3–specific DNAzyme. J. Allergy Clin. Immunol. 2008, 121, 910–916. [Google Scholar] [CrossRef]
- Krug, N.; Hohlfeld, J.M.; Kirsten, A.M.; Kornmann, O.; Beeh, K.M.; Kappeler, D.; Korn, S.; Ignatenko, S.; Timmer, W.; Rogon, C.; et al. Allergen-induced asthmatic responses modified by a GATA3-specific DNAzyme. N. Engl. J. Med. 2015, 372, 1987–1995. [Google Scholar] [CrossRef] [Green Version]
- Santiago, F.S.; Lowe, H.C.; Kavurma, M.M.; Chesterman, C.N.; Baker, A.; Atkins, D.G.; Khachigian, L.M. New DNA enzyme targeting Egr-1 mRNA inhibits vascular smooth muscle proliferation and regrowth after injury. Nat. Med. 1999, 5, 1264–1269. [Google Scholar] [CrossRef]
- Cairns, M.J.; Hopkins, T.M.; Witherington, C.; Wang, L.; Sun, L.Q. Target site selection for an RNA-cleaving catalytic DNA. Nat. Biotechnol. 1999, 17, 480–486. [Google Scholar] [CrossRef]
- Yen, L.; Strittmatter, S.M.; Kalb, R.G. Sequence-specific cleavage of Huntingtin mRNA by catalytic DNA. Ann. Neurol. 1999, 46, 366–373. [Google Scholar] [CrossRef]
- Kurreck, J.; Bieber, B.; Jahnel, R.; Erdmann, V.A. Comparative study of DNA enzymes and ribozymes against the same full-length messenger RNA of the vanilloid receptor subtype I. J. Biol. Chem. 2002, 277, 7099–7107. [Google Scholar] [CrossRef] [Green Version]
- Cho, E.A.; Moloney, F.J.; Cai, H.; Au-Yeung, A.; China, C.; Scolyer, R.A.; Yosufi, B.; Raftery, M.J.; Deng, J.Z.; Morton, S.W.; et al. Safety and tolerability of an intratumorally injected DNAzyme, Dz13, in patients with nodular basal-cell carcinoma: A phase 1 first-in-human trial (DISCOVER). Lancet 2013, 381, 1835–1843. [Google Scholar] [CrossRef] [Green Version]
- Fokina, A.A.; Meschaninova, M.I.; Durfort, T.; Venyaminova, A.G.; François, J.C. Targeting insulin-like growth factor I with 10–23 DNAzymes: 2′-O-methyl modifications in the catalytic core enhance mRNA cleavage. Biochemistry 2012, 51, 2181–2191. [Google Scholar] [CrossRef]
- Young, D.D.; Lively, M.O.; Deiters, A. Activation and deactivation of DNAzyme and antisense function with light for the photochemical regulation of gene expression in mammalian cells. J. Am. Chem. Soc. 2010, 132, 6183–6193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sidorov, A.V.; Grasby, J.A.; Williams, D.M. Sequence-specific cleavage of RNA in the absence of divalent metal ions by a DNAzyme incorporating imidazolyl and amino functionalities. Nucleic Acids Res. 2004, 32, 1591–1601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kasprowicz, A.; Stokowa-Sołtys, K.; Jeżowska-Bojczuk, M.; Wrzesiński, J.; Ciesiołka, J. Characterization of highly efficient RNA-cleaving DNAzymes that function at acidic pH with no divalent metal-ion cofactors. ChemistryOpen 2017, 6, 46–56. [Google Scholar] [CrossRef] [Green Version]
- Hollenstein, M.; Hipolito, C.J.; Lam, C.H.; Perrin, D.M. A self-cleaving DNA enzyme modified with amines, guanidines and imidazoles operates independently of divalent metal cations (M2+). Nucleic Acids Res. 2009, 37, 1638–1649. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lermer, L.; Roupioz, Y.; Ting, R.; Perrin, D.M. Toward an RNaseA mimic: A DNAzyme with imidazoles and cationic amines. J. Am. Chem. Soc. 2002, 124, 9960–9961. [Google Scholar] [CrossRef]
- Zhang, X.; Xu, Y.; Ling, H.; Hattori, T. Inhibition of infection of incoming HIV-1 virus by RNA-cleaving DNA enzyme. FEBS Lett. 1999, 458, 151–156. [Google Scholar] [CrossRef] [Green Version]
- Basu, S.; Sriram, B.; Goila, R.; Banerjea, A.C. Targeted cleavage of HIV-1 coreceptor-CXCR-4 by RNA-cleaving DNA-enzyme: Inhibition of coreceptor function. Antivir. Res. 2000, 46, 125–134. [Google Scholar] [CrossRef]
- Goila, R.; Banerjea, A.C. Sequence specific cleavage of the HIV-1 coreceptor CCR5 gene by a hammer-head ribozyme and a DNA-enzyme: Inhibition of the coreceptor function by DNA-enzyme. FEBS Lett. 1998, 436, 233–238. [Google Scholar] [CrossRef] [Green Version]
- Jakobsen, M.R.; Haasnoot, J.; Wengel, J.; Berkhout, B.; Kjems, J. Efficient inhibition of HIV-1 expression by LNA modified antisense oligonucleotides and DNAzymes targeted to functionally selected binding sites. Retrovirology 2007, 4, 29. [Google Scholar] [CrossRef] [Green Version]
- Khachigian, L.M. Deoxyribozymes as catalytic nanotherapeutic agents. Cancer Res. 2019, 79, 879–888. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Breaker, R.R.; Emilsson, G.M.; Lazarev, D.; Nakamura, S.; Puskarz, I.J.; Roth, A.; Sudarsan, N. A common speed limit for RNA-cleaving ribozymes and deoxyribozymes. RNA 2003, 9, 949–957. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santoro, S.W.; Joyce, G.F. Mechanism and utility of an RNA-cleaving DNA enzyme. Biochemistry 1998, 37, 13330–13342. [Google Scholar] [CrossRef]
- Nowakowski, J.; Shim, P.J.; Prasad, G.S.; Stout, C.D.; Joyce, G.F. Crystal structure of an 82-nucleotide RNA–DNA complex formed by the 10-23 DNA enzyme. Nat. Struct. Biol. 1999, 6, 151–156. [Google Scholar] [PubMed]
- Nowakowski, J.; Shim, P.J.; Joyce, G.F.; Stout, C.D. Crystallization of the 10-23 DNA enzyme using a combinatorial screen of paired oligonucleotides. Acta Crystallogr. Sect. D Biol. Crystallogr. 1999, 55, 1885–1892. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kondo, J.; Takénaka, A. Crystallization of the most active RNA-cleaving deoxyribozyme. Nucleic Acids Symp. Ser. 2000, 44, 201–202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dolot, R.; Sobczak, M.; Mikołajczyk, B.; Nawrot, B. Synthesis, crystallization and preliminary crystallographic analysis of a 52-nucleotide DNA/2′-OMe-RNA oligomer mimicking 10–23 DNAzyme in the complex with a substrate. Nucleosides Nucleotides Nucleic Acids 2017, 36, 292–301. [Google Scholar] [CrossRef]
- Taylor, G.L. Introduction to phasing. Acta Crystallogr. Sect. D 2010, 66, 325–338. [Google Scholar] [CrossRef]
- Keel, A.Y.; Rambo, R.P.; Batey, R.T.; Kieft, J.S. A general strategy to solve the phase problem in RNA crystallography. Structure 2007, 15, 761–772. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Yu, X.; Chen, Y.; Zhang, J.; Wu, B.; Zheng, L.; Haruehanroengra, P.; Wang, R.; Li, S.; Lin, J.; et al. Crystal structure of an RNA-cleaving DNAzyme. Nat. Commun. 2017, 8, 1–10. [Google Scholar]
- Ferré-D’Amaré, A.R.; Zhou, K.; Doudna, J.A. Crystal structure of a hepatitis delta virus ribozyme. Nature 1998, 395, 567–574. [Google Scholar] [CrossRef] [PubMed]
- Ponce-Salvatierra, A.; Wawrzyniak-Turek, K.; Steuerwald, U.; Höbartner, C.; Pena, V. Crystal structure of a DNA catalyst. Nature 2016, 529, 231–234. [Google Scholar] [CrossRef]
- Rosenbach, H.; Victor, J.; Borggräfe, J.; Biehl, R.; Steger, G.; Etzkorn, M.; Span, I. Expanding crystallization tools for nucleic acid complexes using U1A protein variants. J. Struct. Biol. 2020, 210, 107480. [Google Scholar] [CrossRef] [PubMed]
- Plashkevych, O.; Chattopadhyaya, J. Structure of 10–23 DNAzyme in complex with the target RNA in silico—A progress report on the mechanism of RNA cleavage by DNA enzyme. Med. Chem. Nucleic Acids 2011, 272–291. [Google Scholar]
- Choe, Y.J.; Han, H.J.; Lee, J.H.; Seo, S.W.; Choe, B.S. Synthesis and NMR studies of the RNA-cleaving DNA enzyme. Bull. Korean Chem. Soc. 2000, 21, 1025–1030. [Google Scholar]
- Tafer, H.; Ameres, S.L.; Obernosterer, G.; Gebeshuber, C.A.; Schroeder, R.; Martinez, J.; Hofacker, I.L. The impact of target site accessibility on the design of effective siRNAs. Nat. Biotechnol. 2008, 26, 578–583. [Google Scholar] [CrossRef] [PubMed]
- Turner, D.; Sugimoto, N.; Freier, S. Thermodynamics and kinetics of base-pairing and of DNA and RNA self-assembly and helix coil transition. In Nucleic Acids, Subvolume c, Physical Data I, Spectroscopic and Kinetic Data; Saenger, W., Ed.; Landolt-Börnstein, Group VII Biophysics; Springer: Berlin/Heidelberg, Germany, 1990; Volume I, pp. 201–212. [Google Scholar]
- Steger, G. Thermal denaturation of double-stranded nucleic acids: Prediction of temperatures critical for gradient gel electrophoresis and polymerase chain reaction. Nucleic Acids Res. 1994, 22, 2760–2768. [Google Scholar] [CrossRef]
- Sugimoto, N.; Nakano, S.i.; Katoh, M.; Matsumura, A.; Nakamuta, H.; Ohmichi, T.; Yoneyama, M.; Sasaki, M. Thermodynamic parameters to predict stability of RNA/DNA hybrid duplexes. Biochemistry 1995, 34, 11211–11216. [Google Scholar] [CrossRef]
- Cairns, M.J.; Hopkins, T.M.; Witherington, C.; Sun, L.Q. The influence of arm length asymmetry and base substitution on the activity of the 10-23 DNA enzyme. Antisense Nucleic Acid Drug Dev. 2000, 10, 323–332. [Google Scholar] [CrossRef]
- Forconi, M.; Herschlag, D. Metal ion-based RNA cleavage as a structural probe. Methods Enzymol. 2009, 468, 91–106. [Google Scholar]
- Yim, T.; Liu, J.; Lu, Y.; Kane, R.; Dordick, J. Highly active and stable DNAzyme-carbon nanotube hybrids. J. Am. Chem. Soc. 2005, 127, 12200–12201. [Google Scholar] [CrossRef] [PubMed]
- Brown, A.; Liu, J.; He, Y.; Lu, Y. Biochemical characterization of a uranyl ion-specific DNAzyme. Chembiochem 2009, 10, 486–492. [Google Scholar] [CrossRef]
- You, M.; Zhu, Z.; Liu, H.; Gulbakan, B.; Han, D.; Wang, R.; Williams, K.; Tan, W. Pyrene-assisted efficient photolysis of disulfide bonds in DNA-based molecular engineering. ACS Appl. Mater. Interfaces 2010, 2, 3601–3605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thorne, R.; Chinnapen, D.F.; Sekhon, G.; Sen, D. A deoxyribozyme, Sero1C, uses light and serotonin to repair diverse pyrimidine dimers in DNA. J. Mol. Biol. 2009, 388, 21–29. [Google Scholar] [CrossRef]
- Cairns, M.J.; King, A.; Sun, L.Q. Optimisation of the 10–23 DNAzyme–substrate pairing interactions enhanced RNA cleavage activity at purine–cytosine target sites. Nucleic Acids Res. 2003, 31, 2883–2889. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, J.; Li, Z.; Yang, Z.; He, J. Studies on the preferred uracil–adenine base pair at the cleavage site of 10–23 DNAzyme by functional group modifications on adenine. Bioorg. Med. Chem. 2015, 23, 4256–4263. [Google Scholar] [CrossRef]
- Asanuma, H.; Hayashi, H.; Zhao, J.; Liang, X.; Yamazawa, A.; Kuramochi, T.; Matsunaga, D.; Aiba, Y.; Kashida, H.; Komiyama, M. Enhancement of RNA cleavage activity of 10–23 DNAzyme by covalently introduced intercalator. Chem. Commun. 2006, 5062–5064. [Google Scholar] [CrossRef]
- Zhu, J.; Li, Z.; Wang, Q.; Liu, Y.; He, J. The contribution of adenines in the catalytic core of 10-23 DNAzyme improved by the 6-amino group modifications. Bioorg. Med. Chem. Lett. 2016, 26, 4462–4465. [Google Scholar] [CrossRef]
- He, J.; Zhang, D.; Wang, Q.; Wei, X.; Cheng, M.; Liu, K. A novel strategy of chemical modification for rate enhancement of 10–23 DNAzyme: A combination of A9 position and 8-aza-7-deaza-2′-deoxyadenosine analogs. Org. Biomol. Chem. 2011, 9, 5728–5736. [Google Scholar] [CrossRef]
- Wang, Q.; Zhang, D.; Liu, Y.; Cheng, M.; He, J.; Liu, K. A structure-activity relationship study for 2′-deoxyadenosine analogs at A9 position in the catalytic core of 10-23 DNAzyme for rate enhancement. Nucleic Acid Ther. 2012, 22, 423–427. [Google Scholar] [CrossRef]
- Vester, B.; Lundberg, L.B.; Sørensen, M.D.; Babu, B.R.; Douthwaite, S.; Wengel, J. LNAzymes: Incorporation of LNA-type monomers into DNAzymes markedly increases RNA cleavage. J. Am. Chem. Soc. 2002, 124, 13682–13683. [Google Scholar] [CrossRef] [PubMed]
- Schubert, S.; Gül, D.C.; Grunert, H.P.; Zeichhardt, H.; Erdmann, V.A.; Kurreck, J. RNA cleaving ‘10-23’ DNAzymes with enhanced stability and activity. Nucleic Acids Res. 2003, 31, 5982–5992. [Google Scholar] [CrossRef] [PubMed]
- Milman, G.; Langridge, R.; Chamberlin, M. The structure of a DNA-RNA hybrid. Proc. Natl. Acad. Sci. USA 1967, 57, 1804. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nawrot, B.; Widera, K.; Wojcik, M.; Rebowska, B.; Nowak, G.; Stec, W.J. Mapping of the functional phosphate groups in the catalytic core of deoxyribozyme 10-23. FEBS J. 2007, 274, 1062–1072. [Google Scholar] [CrossRef] [PubMed]
- Räz, M.H.; Hollenstein, M. Probing the effect of minor groove interactions on the catalytic efficiency of DNAzymes 8-17 and 10-23. Mol. BioSyst. 2015, 11, 1454–1461. [Google Scholar] [CrossRef] [Green Version]
- Smuga, D.; Majchrzak, K.; Sochacka, E.; Nawrot, B. RNA-cleaving 10-23 deoxyribozyme with a single amino acid-like functionality operates without metal ion cofactors. New J. Chem. 2010, 34, 934–948. [Google Scholar] [CrossRef]
- Li, Z.; Liu, Y.; Liu, G.; Zhu, J.; Zheng, Z.; Zhou, Y.; He, J. Position-specific modification with imidazolyl group on 10–23 DNAzyme realized catalytic activity enhancement. Bioorg. Med. Chem. 2014, 22, 4010–4017. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Cao, L.; Chiuman, W.; Li, Y.; Xi, Z. Probing the function of nucleotides in the catalytic cores of the 8-17 and 10-23 DNAzymes by abasic nucleotide and C3 spacer substitutions. Biochemistry 2010, 49, 7553–7562. [Google Scholar] [CrossRef]
- Zaborowska, Z.; Fürste, J.P.; Erdmann, V.A.; Kurreck, J. Sequence requirements in the catalytic core of the “10-23” DNA enzyme. J. Biol. Chem. 2002, 277, 40617–40622. [Google Scholar] [CrossRef] [Green Version]
- Robaldo, L.; Izzo, F.; Dellafiore, M.; Proietti, C.; Elizalde, P.V.; Montserrat, J.M.; Iribarren, A.M. Influence of conformationally restricted pyrimidines on the activity of 10-23 DNAzymes. Bioorg. Med. Chem. 2012, 20, 2581–2586. [Google Scholar] [CrossRef]
- Zaborowska, Z.; Schubert, S.; Kurreck, J.; Erdmann, V.A. Deletion analysis in the catalytic region of the 10-23 DNA enzyme. FEBS Lett. 2005, 579, 554–558. [Google Scholar] [CrossRef] [Green Version]
- Okumoto, Y.; Sugimoto, N. Effects of metal ions and catalytic loop sequences on the complex formation of a deoxyribozyme and its RNA substrate. J. Inorg. Biochem. 2000, 82, 189–195. [Google Scholar] [CrossRef]
- Sugimoto, N.; Okumoto, Y.; Ohmichi, T. Effect of metal ions and sequence of deoxyribozymes on their RNA cleavage activity. J. Chem. Soc. Perkin Trans. 1999, 2, 1381–1386. [Google Scholar]
- Arnott, S.; Chandrasekaran, R.; Millane, R.; Park, H.S. DNA-RNA hybrid secondary structures. J. Mol. Biol. 1986, 188, 631–640. [Google Scholar] [CrossRef]
- Arnott, S.; Chandrasekaran, R.; Millane, R.; Park, H.S. RNA-RNA, DNA-DNA, and DNA-RNA Polymorphism. Biophys. J. 1986, 49, 3–5. [Google Scholar] [CrossRef] [Green Version]
- Pearson, R.G. Hard and soft acids and bases, HSAB, part 1: Fundamental principles. J. Chem. Educ. 1968, 45, 581. [Google Scholar] [CrossRef]
- Keiper, S.; Vyle, J.S. Reversible photocontrol of deoxyribozyme-catalyzed RNA cleavage under multiple-turnover conditions. Angew. Chem. Int. Ed. 2006, 45, 3306–3309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, P.; Du, S.; Li, Y.; He, J. Studies on the two thymine residues in the catalytic core of 10-23 DNAzyme: The impact on the catalysis of their 5-substituted functional groups. Molecules 2017, 22, 1011. [Google Scholar] [CrossRef] [Green Version]
- Robaldo, L.; Montserrat, J.M.; Iribarren, A.M. 10-23 DNAzyme modified with (2′ R)- (2′ S)-2′ Catalytic Core. Bioorg. Med. Chem. Lett. 2010, 20, 4367–4370. [Google Scholar] [CrossRef]
- Pley, H.W.; Flaherty, K.M.; McKay, D.B. Model for an RNA tertiary interaction from the structure of an intermolecular complex between a GAAA tetraloop and an RNA helix. Nature 1994, 372, 111–113. [Google Scholar] [CrossRef]
- Brown, I. What factors determine cation coordination numbers? Acta Crystallogr. Sect. B 1988, 44, 545–553. [Google Scholar] [CrossRef]
- Hanna, R.; Doudna, J.A. Metal ions in ribozyme folding and catalysis. Curr. Opin. Chem. Biol. 2000, 4, 166–170. [Google Scholar] [CrossRef]
- Donghi, D.; Schnabl, J. Multiple Roles of Metal Ions in Large Ribozymes. In Structural and Catalytic Roles of Metal Ions in RNA; The Royal Society of Chemistry: London, UK, 2011; Volume 9, pp. 197–234. [Google Scholar]
- Fedor, M.J. The role of metal ions in RNA catalysis. Curr. Opin. Struct. Biol. 2002, 12, 289–295. [Google Scholar] [CrossRef]
- Lilley, D.M. Structure, folding and mechanisms of ribozymes. Curr. Opin. Struct. Biol. 2005, 15, 313–323. [Google Scholar] [CrossRef] [PubMed]
- Doherty, E.A.; Doudna, J.A. Ribozyme structures and mechanisms. Annu. Rev. Biophys. Biomol. Struct. 2001, 30, 457–475. [Google Scholar] [CrossRef]
- Sigel, R.K.; Pyle, A.M. Alternative roles for metal ions in enzyme catalysis and the implications for ribozyme chemistry. Chem. Rev. 2007, 107, 97–113. [Google Scholar] [CrossRef] [Green Version]
- Schnabl, J.; Sigel, R.K. Controlling ribozyme activity by metal ions. Curr. Opin. Chem. Biol. 2010, 14, 269–275. [Google Scholar] [CrossRef] [Green Version]
- Pechlaner, M.; Sigel, R.K. Characterization of metal ion-nucleic acid interactions in solution. In Interplay between Metal Ions and Nucleic Acids; Springer: Berlin/Heidelberg, Germany, 2012; pp. 1–42. [Google Scholar]
- Scott, W.G.; Murray, J.B.; Arnold, J.R.; Stoddard, B.L.; Klug, A. Capturing the structure of a catalytic RNA intermediate: The hammerhead ribozyme. Science 1996, 274, 2065–2069. [Google Scholar] [CrossRef] [Green Version]
- Murray, J.B.; Szöke, H.; Szöke, A.; Scott, W.G. Capture and visualization of a catalytic RNA enzyme-product complex using crystal lattice trapping and X-ray holographic reconstruction. Mol. Cell 2000, 5, 279–287. [Google Scholar] [CrossRef]
- Chi, Y.I.; Martick, M.; Lares, M.; Kim, R.; Scott, W.G.; Kim, S.H. Capturing hammerhead ribozyme structures in action by modulating general base catalysis. PLoS Biol. 2008, 6. [Google Scholar] [CrossRef]
- Martick, M.; Lee, T.S.; York, D.M.; Scott, W.G. Solvent structure and hammerhead ribozyme catalysis. Chem. Biol. 2008, 15, 332–342. [Google Scholar] [CrossRef] [Green Version]
- Lippert, B. Multiplicity of metal ion binding patterns to nucleobases. Coord. Chem. Rev. 2000, 200, 487–516. [Google Scholar] [CrossRef]
- He, Q.C.; Zhou, J.M.; Zhou, D.M.; Nakamatsu, Y.; Baba, T.; Taira, K. Comparison of metal-ion-dependent cleavages of RNA by a DNA enzyme and a hammerhead ribozyme. Biomacromolecules 2002, 3, 69–83. [Google Scholar] [CrossRef] [PubMed]
- Cieslak, M.; Szymanski, J.; Adamiak, R.W.; Cierniewski, C.S. Structural rearrangements of the 10-23 DNAzyme to β3 integrin subunit mRNA induced by cations and their relations to the catalytic activity. J. Biol. Chem. 2003, 278, 47987–47996. [Google Scholar] [CrossRef] [Green Version]
- Curtis, E.A.; Bartel, D.P. The hammerhead cleavage reaction in monovalent cations. RNA 2001, 7, 546–552. [Google Scholar] [CrossRef] [Green Version]
- Murray, J.B.; Seyhan, A.A.; Walter, N.G.; Burke, J.M.; Scott, W.G. The hammerhead, hairpin and VS ribozymes are catalytically proficient in monovalent cations alone. Chem. Biol. 1998, 5, 587–595. [Google Scholar] [CrossRef] [Green Version]
- Kenward, M.; Dorfman, K.D. Coarse-grained brownian dynamics simulations of the 10-23 dnazyme. Biophys. J. 2009, 97, 2785–2793. [Google Scholar] [CrossRef] [Green Version]
- Soukup, G.A.; Breaker, R.R. Relationship between internucleotide linkage geometry and the stability of RNA. RNA 1999, 5, 1308–1325. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Breaker, R.R. Kinetics of RNA degradation by specific base catalysis of transesterification involving the 2′-hydroxyl group. J. Am. Chem. Soc. 1999, 121, 5364–5372. [Google Scholar] [CrossRef]
- Nawrot, B.; Widera, K.; Sobczak, M.; Wojcik, M.; Stec, W.J. Effect of RP and SPPhosphorothioate Substitution at the Scissile Site on the Cleavage Activity of Deoxyribozyme 10-23. Curr. Org. Chem. 2008, 12, 1004–1009. [Google Scholar] [CrossRef]
- Emilsson, G.M.; Nakamura, S.; Roth, A.; Breaker, R.R. Ribozyme speed limits. RNA 2003, 9, 907–918. [Google Scholar] [CrossRef] [Green Version]
- Zhou, D.M.; He, Q.C.; Zhou, J.M.; Taira, K. Explanation by a putative triester-like mechanism for the thio effects and Mn2+ rescues in reactions catalyzed by a hammerhead ribozyme. FEBS Lett. 1998, 431, 154–160. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Zhu, J.; He, J. Conformational studies of 10-23 DNAzyme in solution through pyrenyl-labeled 2′-deoxyadenosine derivatives. Org. Biomol. Chem. 2016, 14, 9846–9858. [Google Scholar] [CrossRef] [PubMed]








| AU | = | GU | ≥ | GC | ≫ | AC |
| RU | RC | |||||
| | | > | | | ||||
| dA | dG | |||||
| RY | RY | AC | AC | |||
| | | > | | | | | ≫ | | | |
| dI | dR | dI | dG | |||
| AU | AU | GU | GU | |||
| | | > | | | | | ≈ | | | |
| dA | DAP | dA | DAP | |||
| RU | RU | RU | ||||
| | | ≈ | | | ≈ | | | ||
| dA | dNA | d(NN)A | ||||
| dN | G a | A a | T a | C a | I a | Abasic b | Spacer b | Deletion c | 2-AP a | DP a | 3-DA d | sG e | NG e |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| G | – | <5 f | <5 f | <5 f | 40 f | 106 g | 100 g | <10 f | – | – | – | – | – |
| G | – | <5 f | <5 f | <5 f | 60 f | 97 g | 88 h | <10 f | – | – | – | – | – |
| C | 10 f | 70 f | 20 f | – | <5 f | 100 g | 103 g | 10 f | – | – | – | – | – |
| T | <5 f | <5 f | – | 5 f | <5 f | 73 h | 75 h | <5 f | – | – | – | – | – |
| A | <5 f | – | 5 f | 55 f | 10 f | 6 h | – | <5 f | – | 105 f | 100 g | – | – |
| G | – | <5 f | <5 f | <5 f | 100 f | x | x | <6 f | – | – | – | ||
| C | 20 f | 75 f | 60 | – | 20 f | 79 h | 61 h | 78 f | – | – | – | – | – |
| T | 90 f | 90 f | – | 75 f | 110 f | 106 g | 109 g | 105 f | – | – | – | – | – |
| A | 40 f | – | 90 f | 10 f | 50 f | 71 h | 69 h | 30 f | – | – | 98 g | – | – |
| C | 60 f | 75 f | 60 f | – | 60 f | 47 h | 58 h | 15 f | – | – | – | – | – |
| A | 55 f | – | 85 f | 30 f | 75 f | 94 h | 94 h | 30 f | – | – | 101 g | – | – |
| A | 35 f | – | 85 f | 75 f | 95 f | 97 h | 101 h | 30 f | – | – | 74 g | – | – |
| C | 5 f | 50 f | 10 f | – | 10 f | x | x | <5 f | – | – | – | – | – |
| G | – | <5 f | <5 f | <5 f | 10 f | x | x | <5 f | 5 f | – | – | – | – |
| A | 70 f | – | 50 f | 10 f | 90 f | 84 h | 86 h | <5 f | – | – | 103 g | – | – |
| Reference | Substrate | Length of Binding Arms | Mg [mM] | NaCl [mM] | RNA | DNAzyme | Duration and Temperature | Output |
|---|---|---|---|---|---|---|---|---|
| [17] | RNA | 7/7 | 10 | 150 | 0.2 µM | 0.5–500 nM | various timepoints, 37 °C | , single and multiple turnover |
| [50] | RNA | variable | 10 | – | 0.04 µM | 0.32 µM | various timepoints, 37 °C | , single turnover |
| [56] | RNA | 9/9 | 10 | – | 0.6 µM | 5 µM | various timepoints, up to 1 h; 37 °C | , single turnover |
| [96] | RNA | 7/7 | 25 | – | 0.06–0.1 µM | 0.5–10 µM | various timepoints, 37 °C | , multiple turnover |
| [60] | DNA/w RNA | 9/9 | 6 (2) | – | 0.02 µM (0.2 µM) | 2 µM (0.02 µM) | various timepoints, 37 °C | , single turnover (multiple turnover) |
| [68] | DNA/w RNA | 9/9 | 0.5–10 | – | 0.02 µM | 2 µM | various timepoints, 37 °C | , single turnover |
| [106] | DNA/w RNA | 9/9 | 20 | – | 0.02 µM | 2 µM | various timepoints, 37 °C | , single turnover |
| [79] | DNA/w RNA | 9/9 | 2.18 | – | 0.02 µM | 2 µM | various timepoints, 37 °C | , single turnover |
| [65] | DNA /w RNA | 8/8 | 3 | 100 | 0.1 µM | 10 µM | various timepoints, 37 °C | , single turnover |
| [103] | DNA/w RNA | 8/8 | 0.02 | 100 | 0.1 µM | 10 µM | various timepoints, 37 °C | , single turnover |
| [73] | RNA | 6/8 | 25 | – | 0.01–1 µM | 0.001 µM | various timepoints, 37 °C | , multiple turnover |
| [80] | RNA | 8/8 | 0.5–10 | – | 0.5 µM | 5 µM | various timepoints, 37 °C | , single turnover |
| [71] | RNA | 8/8 | 0.5–10 | – | 0.5 µM | 0.5 µM | various timepoints, 37 °C | , single turnover |
| [1] | RNA | 8/8 | variable | variable | variable | variable | various timepoints, 37 °C | and , single and multiple turnover |
| [33] | RNA | 8/8 | 2 | 150 | variable | variable | various timepoints, 37 °C | and , single and multiple turnover |
| [63] | RNA | 9/9 | 10 | 100 | 1 µM | 0.01 µM | 20 min, 37 °C | initial velocity, multiple turnover |
| [74] | RNA | 8/6 | 25 | – | 5 nM (tenfold excess) | 1 µM (n.a.) | various timepoints, 37 °C | Y (), single turnover (multiple turnover) |
| [11] | RNA | 9/9 | 0.1-1 | 100 | 0.1 µM | 0.1 µM | various timepoints, 37 °C | , single turnover |
| [69] | DNA/w RNA | 9/9 | 500 | 200 | 0.005 µM | 0.5 µM | various timepoints, 37 °C | Y and , single turnover |
| [61] | DNA/w RNA | 9/9 | 2 | – | 0.02 µM | 2 µM | various timepoints, – | Y and , single turnover |
| [70] | RNA (RNA) | 9/9 | 10 | – | 0.1 µM | 1 µM | 20 min, 37 °C | Y, single turnover |
| [72] | RNA (RNA) | 9/9 (9/9) | 10 (10) | – (–) | 1 pmol (0.01–1 µM) | 10 pmol (0.001 µM) | 20 min (various timepoints), 37 °C | Y (, multiple turnover) |
| [57] | DNA/w RNA | 9/9 | 2 | – | n.a. | 100-fold excess | Various timepoints, 37 °C | , single turnover |
| [59] | DNA/w RNA | 9/9 | 2 | – | 0.002 µM | 2 µM | various timepoints, 37 °C | , single turnover |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rosenbach, H.; Victor, J.; Etzkorn, M.; Steger, G.; Riesner, D.; Span, I. Molecular Features and Metal Ions That Influence 10-23 DNAzyme Activity. Molecules 2020, 25, 3100. https://doi.org/10.3390/molecules25133100
Rosenbach H, Victor J, Etzkorn M, Steger G, Riesner D, Span I. Molecular Features and Metal Ions That Influence 10-23 DNAzyme Activity. Molecules. 2020; 25(13):3100. https://doi.org/10.3390/molecules25133100
Chicago/Turabian StyleRosenbach, Hannah, Julian Victor, Manuel Etzkorn, Gerhard Steger, Detlev Riesner, and Ingrid Span. 2020. "Molecular Features and Metal Ions That Influence 10-23 DNAzyme Activity" Molecules 25, no. 13: 3100. https://doi.org/10.3390/molecules25133100