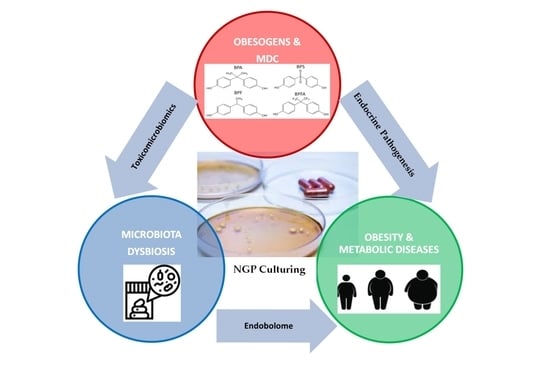
Next Generation Probiotics for Neutralizing Obesogenic Effects: Taxa Culturing Searching Strategies
Abstract
:1. Introduction
1.1. Microbiota Gut Dysbiosis
1.2. Traditional Probiotics vs. NGP in Obesity-Related Interventions and Treatments
| Lactobacillus Strains [15] | Study Design, Target Species | Reference Study |
|---|---|---|
| L. bulgaricus Nutricion Medica® | ICT—Human | [16] |
| L. casei Shirota | ICT—Human | [17] |
| L. gasseri BNR17 | ICT—Human | [18] |
| L. reuteri V3401 | ICT—Human | [19] |
| L. rhamnosus CGMCC1.3724 | ICT—Human | [20] |
| L. acidophilus NS1 | PCS—Mice | [21] |
| L. johnsonii JNU3402 | PCS—Mice | [22] |
| L. plantarum Ln4 | PCS—Mice | [23] |
| L.curvatus HY7601 | PCS—Mice | [24] |
| L. fermentum CQPC07 | PCS—Mice | [25] |
| Bifidobacterium strains | Study design, Target Species, | Reference study |
| B. animalis subsp. lactis 420 | ICT—Human | [26] |
| B. breve B-3 | ICT—Human | [27] |
| B. infantis DSM24737 (VSL#3) | ICT—Human | [28] |
| B. lactis HN019 | ICT—Human | [29] |
| B. longum APC1472 | ICT–Human/PCS–Mice | [30] |
| B. adolescentis | PCS—Mice | [31] |
| B. bifidum BGN4 | PCS—Mice | [32] |
| Bacillus, Enterococcus, Streptococcus strains | Study design, Target Species, | Reference study |
| Bacillus coagulans Unique IS2 | ICT—Human | [33] |
| Bacillus amyloliquefaciens SC06 | PCS—Mice | [34] |
| Bacillus spp. | PCS—Mice | [35] |
| Enterococcus faecium R0026 | PCS—Mice | [36] |
| Enterococcus faecalis AG5 | PCS—Rats | [37] |
| Streptococcus thermophiles MN-ZLW-002 | PCS—Mice | [38] |
| Saccharomyces strains | Study design, Target Species, | Reference study |
| S. boulardii Biocodex | PCS–Mice | [39] |
| S. cerevisiae SFBE | PCS–Rats | [40] |
2. Information and Criteria for Searching and Culturing Next-Generation Probiotics
2.1. Target Diseases, Microbiome Variability Composition, Biomarkers and Clinical Traits
2.1.1. Obesity, Metabolic, and Endocrine Diseases: Variability of Microbiota Composition
2.1.2. Nutrition and Diets, Dietary Exposure to Obesogens, and Microbiome Interactions
2.2. Culturing and Isolation of NGP through Combined Methodologies
2.3. Standardize Parameters When Using NGP in Clinical Studies
2.4. Whole Genome Sequencing, Next-Generation Sequencing, and Bioinformatics Analyses
2.5. Omics Data Integration: Big Data and Host Clinical Responses
2.6. Safety Assessment, Regulatory Frameworks, and Market Labeling
| Country | Category | Regulatory Framework | Claims | Reference |
|---|---|---|---|---|
| USA | Drugs, nutraceuticals | FDA | Health claims Nutrient claims Structure claims GRAS | [145,146] |
| Dietary supplements | DSHEA | Probiotics considered as foods | ||
| Biological product | FDA (BLA) | Probiotics as a reference product, biosimilar product, or an interchangeable product; solely to be used for medical therapeutic purpose | ||
| Life biotherapeutic agent | FDA | Probiotics as a biological product that contains live organisms and is applicable to the prevention, treatment, or cure of a disease or condition; recombinant life biotherapeutic agent | ||
| Medical Food | FDA/DSHA | Probiotics specially formulated to be intended for dietary management under supervision; medical foods are exempt from the labeling requirements for nutrient content and health claims | ||
| China | Functional foods | SFDA | Conventional foods mark (the presence of a specific ingredient in the label of regular foodstuffs) Healthy foods (the presence of health function) | [147] |
| Europe | Functional Food and nutraceuticals | EFSA (FUFOSE) | Health claims, nutrition claims QPS | [143,144,148] |
| Life biotherapeutic products | EMA | Probiotics as medicinal products containing live microorganisms for human use | ||
| Japan | Functional foods and nutraceuticals | MHLW, FOSHU | Foods with functional claims Foods with nutrient functional claims | [149,150] |
| Canada | Natural health products | FDA (CFIA) | Nutrient content claims Health claims | [151] |
3. Discussion
4. Conclusions
- Culturing of microorganisms from microbiota is the key activity to obtain NGP from healthy individuals, mainly through isolating those microorganisms identified as differentially decreased in the target disease or abundant in healthy microbiota, focusing on candidatus species from metagenomics studies.
- Screening and selection of the potential NGP in a target-disease population by using in vitro models before clinical interventions.
- Harmonization on performing exhaustive pre-analysis and post-intervention of individual microbiota composition through representative and validated methodologies (e.g., V3–V4 and Illumina MiSeq technology) is needed before administering NGP.
- There is a need to standardize bioinformatics and database tools for specifically designing analysis of large and universal microbiome datasets.
- NGP single strains or taxa consortium should have attributable documented benefits and their safety confirmation statements.
- Effective doses and well-defined patterns of administration of NGP should become factors for aligning intervention doses since the beginning of clinical translation.
- International guidelines on NGP and microbiota investigations for targeting obesity-related diseases prevention or treatments are needed. This will allow for more meaningful effect comparisons of harmonized and valuable studies, facilitating more robust meta-analysis.
- Data reuse and availability of open access interventional clinical trials data will contribute to obtaining significant association of clinical outcomes.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| MDC | Microbiota-disrupting chemicals |
| NGP | Next-generation probiotics |
| GIT | Gastrointestinal tract |
| PCOS | Polycystic ovary syndrome |
| FAO | Food and Agriculture Organization of the United Nations |
| WHO | World Health Organization |
| ICT | Interventional clinical trials |
| PCS | Preclinical studies |
| DC | Dendritic cells |
| IL | Interleukin |
| LPS | Lipopolysaccharide |
| TLR2 | Toll-like receptor 2 |
| TNF | Tumor necrosis factor |
| WGS | Whole genome sequencing |
| NGS | New-generation sequencing |
| AN | Anorexia nervosa |
| HC | Healthy control |
| HL | Hyperlipidemia |
| HT | Hypertension |
| LH | Lean healthy |
| MetS | Metabolic syndrome |
| MHNO | Metabolically healthy non-obese |
| MHO | Metabolically healthy obese |
| MUNO | Metabolically unhealthy non-obese |
| MUO | Metabolically unhealthy obese |
| NAFLD | Non-alcoholic fatty liver disease |
| NASH | Non-alcoholic steatohepatitis |
| OB | Obese |
| OBH | Obese healthy |
| OBT2D | Obese type 2 diabetes |
| OW | Overweight |
| RISK1 | Patients with only one disease |
| RISK2 | Patients with two disease |
| RISK3 | Patients with three disease |
| SS | Simple steatosis |
| T1D | Type 1 diabetes |
| T2D | Type 2 diabetes |
| TSNO | Tsumura Suzuki obese diabetes mice |
| TSOD | Tsumura Suzuki non obesity mice |
| BPA | Bisphenol A |
| BPS | Bisphenol S |
| YCFA | Yeast-extract-casein hydrolysate-fatty acids |
| GAM | Gifu anaerobic medium |
| BHI | Brain–heart infusion |
| EMB | Eosin methylene blue |
| LBS | Lactobacillus selection |
| GMM | Gut microbiota medium |
| MRS | Man, Rogosa, and Sharpe |
| RNA | Ribonucleic acid |
| rRNA | Ribosomal ribonucleic acid |
| DNA | Deoxyribonucleic acid |
| OTU | Operational taxonomic unit |
| EU | European Union |
| Ph. Eur. | European Pharmacopoeia |
| US | United States |
| GRAS | Generally recognized as safe |
| FDA | Food and Drug Administration |
| EFSA | European Food Safety Authority |
| QPS | Qualified presumption of safety |
| EMA | European Medicines Agency |
| MHLW | Ministry of Health and Welfare |
| FOSHU | Food for specified health use |
| FUFOSE | Functional food science in Europe |
| SFDA | State Food and Drug Administration |
| DSHEA | Dietary Supplement Health and Education Act |
| BLA | Biologic license application |
| CFIA | The Canadian Food Inspection Agency |
| GMP | Good manufacturing practice |
| CFU | Colony-forming units |
| OA | Open access |
References
- Katsi, V.; Didagelos, M.; Skevofilax, S.; Armenis, I.; Kartalis, A.; Vlachopoulos, C.; Karvounis, H.; Tousoulis, D. GUT Microbiome-GUT dysbiosis-arterial hypertension: New horizons. Curr. Hypertens. Rev. 2019, 15, 40–46. [Google Scholar] [CrossRef] [PubMed]
- Daliri, E.B.-M.; Lee, B.H. New perspectives on probiotics in health and disease. Food Sci. Hum. Wellness 2015, 4, 56–65. [Google Scholar] [CrossRef] [Green Version]
- Ohkusa, T.; Koido, S.; Nishikawa, Y.; Sato, N. Gut microbiota and chronic constipation: A review and update. Front. Med. 2019, 6, 19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsuoka, K.; Kanai, T. The gut microbiota and inflammatory bowel disease. Semin. Immunopathol. 2015, 37, 47–55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- dos Santos, C.; Ferreira, K.; Midori, P.; Fábio, L.; Darrieux, M.; Manzano, T. Childhood obesity and Firmicutes/Bacteroidetes ratio in the gut microbiota: A systematic review. Child. Obes. 2018, 14, 501–509. [Google Scholar] [CrossRef]
- Liang, Y.; Ming, Q.; Liang, J.; Zhang, Y.; Zhang, H.; Shen, T. Gut microbiota dysbiosis in Polycystic Ovary Syndrome: Association with obesity-a preliminary report. Can. J. Physiol. Pharm. 2020, 98, 803–809. [Google Scholar] [CrossRef] [PubMed]
- Meng, C.; Bai, C.; Brown, T.D.; Hood, L.E.; Tian, Q. Human gut microbiota and gastrointestinal cancer. Genom. Proteom. Bioinform. 2018, 16, 33–49. [Google Scholar] [CrossRef]
- Bear, T.L.K.; Dalziel, J.E.; Coad, J.; Roy, N.C.; Butts, C.A.; Gopal, P.K. The role of the gut microbiota in dietary interventions for depression and anxiety. Adv. Nutr. 2020, 11, 890–907. [Google Scholar] [CrossRef] [Green Version]
- Food and Agriculture Organization; World Health Organization Expert Consultation. Evaluation of Health and Nutritional Properties of Powder Milk and Live Lactic Acid Bacteria. Córdoba, Argentina: Food and Agriculture Organization of the United Nations and World Health Organization. 2001. Available online: ftp://ftp.fao.org/es/esn/food/probio_report_en.pdf (accessed on 8 February 2021).
- Chang, C.-J.; Lin, T.-L.; Tsai, Y.-L.; Wu, T.-R.; Lai, W.-F.; Lu, C.-C.; Lai, H.-C. Next Generation Probiotics in disease amelioration. J. Food Drug Anal. 2019, 27, 615–622. [Google Scholar] [CrossRef]
- Tan, H.; Zhai, Q.; Chen, W. Investigations of Bacteroides spp. towards next-generation probiotics. Food Res. Int. 2019, 116, 637–644. [Google Scholar] [CrossRef]
- Azaïs-Braesco, V.; Bresson, J.L.; Guarner, F.; Corthier, G. Not All Lactic Acid Bacteria Are Probiotics, but Some Are. Br. J. Nutr. 2010, 103, 1079–1081. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rijkers, G.T.; de Vos, W.M.; Brummer, R.-J.; Morelli, L.; Corthier, G.; Marteau, P. Health Benefits and Health Claims of Probiotics: Bridging Science and Marketing. Br. J. Nutr. 2011, 106, 1291–1296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Toole, P.W.; Marchesi, J.R.; Hill, C. Next-Generation Probiotics: The spectrum from probiotics to live biotherapeutics. Nat. Microbiol. 2017, 2, 17057. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Wittouck, S.; Salvetti, E.; Franz, C.; Harris, H.; Mattarelli, P.; O’Toole, P.; Pot, B.; Vandamme, P.; Walter, J.; et al. A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 2020, 70, 2782–2858. [Google Scholar] [CrossRef] [PubMed]
- Aller, R.; De Luis, D.A.; Izaola, O.; Conde, R.; Gonzalez, S.M.; Primo, D.; De La Fuente, B.; Gonzalez, J. Effect of a probiotic on liver aminotransferases in nonalcoholic fatty liver disease patients: A double blind randomized clinical trial. Eur. Rev. Med. Pharm. Sci. 2011, 15, 1090–1095. [Google Scholar]
- Nagata, S.; Chiba, Y.; Wang, C.; Yamashiro, Y. The effects of the Lactobacillus casei strain on obesity in children: A pilot study. Benef. Microbes 2017, 8, 535–543. [Google Scholar] [CrossRef]
- Kim, J.; Yun, J.M.; Kim, M.K.; Kwon, O.; Cho, B. Lactobacillus gasseri BNR17 supplementation reduces the visceral fat accumulation and waist circumference in obese adults: A randomized, double-blind, placebo-controlled trial. J. Med. Food 2018, 21, 454–461. [Google Scholar] [CrossRef]
- Tenorio-Jiménez, C.; Martínez-Ramírez, M.J.; Tercero-Lozano, M.; Arraiza-Irigoyen, C.; Del Castillo-Codes, I.; Olza, J.; Plaza-Díaz, J.; Fontana, L.; Migueles, J.H.; Olivares, M.; et al. Evaluation of the effect of Lactobacillus Reuteri V3401 on biomarkers of inflammation, cardiovascular risk and liver steatosis in obese adults with metabolic syndrome: A randomized clinical trial (PROSIR). BMC Complement. Altern. Med. 2018, 18, 306. [Google Scholar] [CrossRef]
- Sánchez, M.; Darimont, C.; Drapeau, V.; Emady-Azar, S.; Lepage, M.; Rezzonico, E.; Ngom-Bru, C.; Berger, B.; Philippe, L.; Ammon-Zuffrey, C.; et al. Effect of Lactobacillus rhamnosus CGMCC1.3724 supplementation on weight loss and maintenance in obese men and women. Br. J. Nutr. 2014, 111, 1507–1519. [Google Scholar] [CrossRef] [Green Version]
- Park, S.-S.; Lee, Y.-J.; Song, S.; Kim, B.; Kang, H.; Oh, S.; Kim, E. Lactobacillus acidophilus NS1 attenuates diet-induced obesity and fatty liver. J. Endocrinol. 2018, 237, 87–100. [Google Scholar] [CrossRef]
- Yang, G.; Hong, E.; Oh, S.; Kim, E. Non-Viable Lactobacillus Johnsonii JNU3402 protects against diet-induced obesity. Foods 2020, 9, 1494. [Google Scholar] [CrossRef]
- Lee, E.; Jung, S.-R.; Lee, S.-Y.; Lee, N.-K.; Paik, H.-D.; Lim, S.-I. Lactobacillus plantarum strain Ln4 attenuates diet-induced obesity, insulin resistance, and changes in hepatic mrna levels associated with glucose and lipid metabolism. Nutrients 2018, 10, 643. [Google Scholar] [CrossRef] [Green Version]
- Park, D.-Y.; Ahn, Y.-T.; Park, S.-H.; Huh, C.-S.; Yoo, S.-R.; Yu, R.; Sung, M.-K.; McGregor, R.A.; Choi, M.-S. Supplementation of Lactobacillus curvatus HY7601 and Lactobacillus plantarum KY1032 in diet-induced obese mice is associated with gut microbial changes and reduction in obesity. PLoS ONE 2013, 8, e59470. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.; Li, X.; Tan, F.; Zhou, X.; Mu, J.; Zhao, X. Lactobacillus fermentum CQPC07 attenuates obesity, inflammation and dyslipidemia by modulating the antioxidant capacity and lipid metabolism in high-fat diet induced obese mice. J. Inflamm. 2021, 18, 5. [Google Scholar] [CrossRef] [PubMed]
- Uusitupa, H.-M.; Rasinkangas, P.; Lehtinen, M.J.; Mäkelä, S.M.; Airaksinen, K.; Anglenius, H.; Ouwehand, A.C.; Maukonen, J. Bifidobacterium animalis Subsp. lactis 420 for metabolic health: Review of the research. Nutrients 2020, 12, 892. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Minami, J.; Iwabuchi, N.; Tanaka, M.; Yamauchi, K.; Xiao, J.-Z.; Abe, F.; Sakane, N. Effects of Bifidobacterium breve B-3 on body fat reductions in pre-obese adults: A randomized, double-blind, placebo-controlled trial. Biosci. Microbiota Food Health 2018, 37, 67–75. [Google Scholar] [CrossRef] [Green Version]
- Jones, R.B.; Alderete, T.L.; Martin, A.A.; Geary, B.A.; Hwang, D.H.; Palmer, S.L.; Goran, M.I. Probiotic supplementation increases obesity with no detectable effects on liver fat or gut microbiota in obese hispanic adolescents: A 16-Week, randomized, placebo-controlled trial. Pediatr. Obes. 2018, 13, 705–714. [Google Scholar] [CrossRef] [PubMed]
- Bernini, L.J.; Simão, A.N.C.; de Souza, C.H.B.; Alfieri, D.F.; Segura, L.G.; Costa, G.N.; Dichi, I. Effect of Bifidobacterium lactis HN019 on inflammatory markers and oxidative stress in subjects with and without the metabolic syndrome. Br. J. Nutr. 2018, 120, 645–652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schellekens, H.; Torres-Fuentes, C.; van de Wouw, M.; Long-Smith, C.M.; Mitchell, A.; Strain, C.; Berding, K.; Bastiaanssen, T.F.S.; Rea, K.; Golubeva, A.V.; et al. Bifidobacterium longum counters the effects of obesity: Partial successful translation from rodent to human. EBioMedicine 2021, 63, 103176. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.-M.; Kim, D.-H. Bifidobacterium adolescentis IM38 ameliorates High-Fat diet-induced colitis in mice by inhibiting nf-κb activation and lipopolysaccharide production by gut microbiota. Nutr. Res. 2017, 41, 86–96. [Google Scholar] [CrossRef]
- Li, Z.; Jin, H.; Oh, S.Y.; Ji, G.E. Anti-Obese effects of two Lactobacilli and two Bifidobacteria on ICR mice fed on a high fat diet. Biochem. Biophys. Res. Commun. 2016, 480, 222–227. [Google Scholar] [CrossRef] [PubMed]
- Sudha, M.R.; Ahire, J.J.; Jayanthi, N.; Tripathi, A.; Nanal, S. Effect of multi-strain probiotic (UB0316) in weight management in overweight/obese adults: A 12-Week Double Blind, Randomised, Placebo-Controlled Study. Benef. Microbes 2019, 10, 855–866. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wu, Y.; Wang, B.; Xu, H.; Mei, X.; Xu, X.; Zhang, X.; Ni, J.; Li, W. Bacillus amyloliquefaciens SC06 protects mice against high-fat diet-induced obesity and liver injury via regulating host metabolism and gut microbiota. Front. Microbiol. 2019, 10, 1161. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.; Kwon, J.; Kim, M.-S.; Park, H.; Ji, Y.; Holzapfel, W.; Hyun, C.-K. Protective Effects of Bacillus probiotics against high-fat diet-induced metabolic disorders in mice. PLoS ONE 2018, 13, e0210120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, J.; Huang, J.; Yin, T.; Lv, H.; Zhang, P.; Li, H. Enterococcus faecium R0026 combined with Bacillus subtilis R0179 prevent obesity-associated hyperlipidaemia and modulate gut microbiota in C57BL/6 Mice. J. Microbiol. Biotechnol 2020, 31, 181–188. [Google Scholar] [CrossRef]
- Mishra, A.K.; Ghosh, A.R. Probiotic Enterococcus faecalis AG5 Mitigated high fat diet induced obesity and produced propionic acid stimulated apoptosis in 3T3-L1 pre-adipocyte. Life Sci. 2020, 261, 118292. [Google Scholar] [CrossRef]
- Kang, X.; Liang, H.; Luo, Y.; Li, Z.; He, F.; Han, X.; Zhang, L. Anti-adipogenesis and metabolism-regulating effects of heat-inactivated Streptococcus thermophilus MN-ZLW-002. Lett. Appl. Microbiol. 2020. [Google Scholar] [CrossRef]
- Everard, A.; Matamoros, S.; Geurts, L.; Delzenne, N.M.; Cani, P.D. Saccharomyces boulardii administration changes gut microbiota and reduces hepatic steatosis, low-grade inflammation, and fat mass in obese and Type 2 Diabetic Db/Db Mice. mBio 2014, 5, e01011–e01014. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Xiao, X.; Dong, Y.; Shi, L.; Xu, T.; Wu, F. The anti-obesity effect of fermented barley extracts with Lactobacillus plantarum Dy-1 and Saccharomyces cerevisiae in diet-induced obese rats. Food Funct. 2017, 8, 1132–1143. [Google Scholar] [CrossRef]
- Depommier, C.; Everard, A.; Druart, C.; Plovier, H.; Van Hul, M.; Vieira-Silva, S.; Falony, G.; Raes, J.; Maiter, D.; Delzenne, N.M.; et al. Supplementation with Akkermansia muciniphila in Overweight and Obese Human Volunteers: A Proof-of-Concept Exploratory Study. Nat. Med. 2019, 25, 1096–1103. [Google Scholar] [CrossRef]
- ClinicalTrials.gov Unique Identifier: NCT04797442: Effect of Akkermansia muciniphila WST01 Strain in Overweight or Obese Patients with Type 2 Diabetes. Available online: https://clinicaltrials.gov/ct2/show/NCT04797442 (accessed on 27 April 2021).
- ClinicalTrials.gov Unique Identifier: NCT04663139: Xla1 Christensenella minuta, Phase I, Randomized, Partially Placebo-controlled Double-blind. Available online: https://clinicaltrials.gov/ct2/show/NCT04663139 (accessed on 27 April 2021).
- ClinicalTrials.gov Unique Identifier: NCT04529473: Efficacy and Safety of 12-weeks Supplementation of Eubacterium hallii on Insulin Sensitivity and Glycaemic Control. Available online: https://clinicaltrials.gov/ct2/show/NCT04529473 (accessed on 27 April 2021).
- Déchelotte, P.; Breton, J.; Trotin-Picolo, C.; Grube, B.; Erlenbeck, C.; Bothe, G.; Fetissov, S.O.; Lambert, G. The probiotic strain H. alvei HA4597¬Æ improves weight loss in overweight subjects under moderate hypocaloric diet: A proof-of-concept, multicenter randomized, double-blind placebo-controlled study. Nutrients 2021, in press. [Google Scholar]
- Beltrán-Barrientos, L.M.; Garcia, H.S.; Reyes-Díaz, R.; Estrada-Montoya, M.C.; Torres-Llanez, M.J.; Hernández-Mendoza, A.; González-Córdova, A.F.; Vallejo-Cordoba, B. Cooperation between Lactococcus lactis NRRL B-50571 and NRRL B-50572 for Aroma Formation in Fermented Milk. Foods 2019, 8, 645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bazi, Z.; Jalili, M.; Hekmatdoost, A. The long term oral regulation of blood glucose in diabetic patients by using of Escherichia coli Nissle 1917 expressing CTB-IGF-1 hybrid protein. Med. Hypotheses 2013, 81, 961–962. [Google Scholar] [CrossRef]
- Everard, A.; Belzer, C.; Geurts, L.; Ouwerkerk, J.P.; Druart, C.; Bindels, L.B.; Guiot, Y.; Derrien, M.; Muccioli, G.G.; Delzenne, N.M.; et al. Cross-Talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl. Acad. Sci. USA 2013, 110, 9066–9071. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cani, P.D.; de Vos, W.M. Next-Generation Beneficial Microbes: The case of Akkermansia muciniphila. Front. Microbiol. 2017, 8, 765. [Google Scholar] [CrossRef]
- Shang, H.; Sun, J.; Chen, Y.Q. Clostridium butyricum CGMCC0313.1 modulates lipid profile, insulin resistance and colon homeostasis in obese mice. PLoS ONE 2016, 11, e0154373. [Google Scholar] [CrossRef]
- Munukka, E.; Rintala, A.; Toivonen, R.; Nylund, M.; Yang, B.; Takanen, A.; Hänninen, A.; Vuopio, J.; Huovinen, P.; Jalkanen, S.; et al. Faecalibacterium prausnitzii treatment improves hepatic health and reduces adipose tissue inflammation in high-fat fed mice. ISME J. 2017, 11, 1667–1679. [Google Scholar] [CrossRef] [Green Version]
- Cano, P.G.; Santacruz, A.; Moya, Á.; Sanz, Y. Bacteroides uniformis CECT 7771 ameliorates metabolic and immunological dysfunction in mice with high-fat-diet induced obesity. PLoS ONE 2012, 7, e41079. [Google Scholar] [CrossRef] [Green Version]
- Wu, T.-R.; Lin, C.-S.; Chang, C.-J.; Lin, T.-L.; Martel, J.; Ko, Y.-F.; Ojcius, D.M.; Lu, C.-C.; Young, J.D.; Lai, H.-C. Gut commensal Parabacteroides goldsteinii plays a predominant role in the anti-obesity effects of polysaccharides isolated from Hirsutella sinensis. Gut 2019, 68, 248–262. [Google Scholar] [CrossRef] [PubMed]
- Goodrich, J.K.; Waters, J.L.; Poole, A.C.; Sutter, J.L.; Koren, O.; Blekhman, R.; Beaumont, M.; Van Treuren, W.; Knight, R.; Bell, J.T.; et al. Human genetics shape the gut microbiome. Cell 2014, 159, 789–799. [Google Scholar] [CrossRef] [Green Version]
- Engels, C.; Ruscheweyh, H.-J.; Beerenwinkel, N.; Lacroix, C.; Schwab, C. The common gut microbe eubacterium hallii also contributes to intestinal propionate formation. Front. Microbiol. 2016, 7, 713. [Google Scholar] [CrossRef] [Green Version]
- Lucas, N.; Legrand, R.; Deroissart, C.; Dominique, M.; Azhar, S.; Le Solliec, M.-A.; Léon, F.; do Rego, J.-C.; Déchelotte, P.; Fetissov, S.O.; et al. Hafnia alvei HA4597 strain reduces food intake and body weight gain and improves body composition, glucose, and lipid metabolism in a mouse model of hyperphagic obesity. Microorganisms 2019, 8, 35. [Google Scholar] [CrossRef] [Green Version]
- Ma, Y.; Liu, J.; Hou, J.; Dong, Y.; Lu, Y.; Jin, L.; Cao, R.; Li, T.; Wu, J. Oral administration of recombinant Lactococcus lactis expressing HSP65 and tandemly repeated P277 reduces the incidence of type i diabetes in non-obese diabetic mice. PLoS ONE 2014, 9, e105701. [Google Scholar] [CrossRef]
- Ma, J.; Li, C.; Wang, J.; Gu, J. Genetically engineered Escherichia coli Nissle 1917 Secreting GLP-1 analog exhibits potential antiobesity effect in high-fat diet-induced obesity mice. Obesity 2020, 28, 315–322. [Google Scholar] [CrossRef] [PubMed]
- Gomes, A.C.; Hoffmann, C.; Mota, J.F. The Human Gut Microbiota: Metabolism and perspective in obesity. Gut Microbes 2018, 9, 308–325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riva, A.; Borgo, F.; Lassandro, C.; Verduci, E.; Morace, G.; Borghi, E.; Berry, D. pediatric obesity is associated with an altered gut microbiota and discordant shifts in Firmicutes populations. Environ. Microbiol. 2017, 19, 95–105. [Google Scholar] [CrossRef] [PubMed]
- Munukka, E.; Wiklund, P.; Pekkala, S.; Völgyi, E.; Xu, L.; Cheng, S.; Lyytikäinen, A.; Marjomäki, V.; Alen, M.; Vaahtovuo, J.; et al. Women with and without metabolic disorder differ in their gut microbiota composition. Obesity 2012, 20, 1082–1087. [Google Scholar] [CrossRef]
- Bervoets, L.; Van Hoorenbeeck, K.; Kortleven, I.; Van Noten, C.; Hens, N.; Vael, C.; Goossens, H.; Desager, K.N.; Vankerckhoven, V. Differences in gut microbiota composition between obese and lean children: A cross-sectional study. Gut Pathog. 2013, 5, 10. [Google Scholar] [CrossRef] [Green Version]
- Santacruz, A.; Collado, M.C.; García-Valdés, L.; Segura, M.T.; Martín-Lagos, J.A.; Anjos, T.; Martí-Romero, M.; Lopez, R.M.; Florido, J.; Campoy, C.; et al. Gut microbiota composition is associated with body weight, weight gain and biochemical parameters in pregnant women. Br. J. Nutr. 2010, 104, 83–92. [Google Scholar] [CrossRef] [Green Version]
- Collado, M.C.; Isolauri, E.; Laitinen, K.; Salminen, S. Distinct composition of gut microbiota during pregnancy in overweight and normal-weight women. Am. J. Clin. Nutr. 2008, 88, 894–899. [Google Scholar] [CrossRef]
- Schwiertz, A.; Taras, D.; Schäfer, K.; Beijer, S.; Bos, N.A.; Donus, C.; Hardt, P.D. Microbiota and SCFA in lean and overweight healthy subjects. Obesity 2010, 18, 190–195. [Google Scholar] [CrossRef]
- Zhong, X.; Harrington, J.M.; Millar, S.R.; Perry, I.J.; O’toole, P.W.; Phillips, C.M. Gut microbiota associations with metabolic health and obesity status in older adults. Nutrients 2020, 12, 2364. [Google Scholar] [CrossRef] [PubMed]
- Jonduo, M.E.; Wawae, L.; Masiria, G.; Suda, W.; Hattori, M.; Takayasu, L.; Abdad, M.Y.; Greenhill, A.R.; Horwood, P.F.; Pomat, W.; et al. Gut microbiota composition in obese and non-obese adult relatives from the highlands of Papua New Guinea. Microbiol. Lett. 2020, 367, 19. [Google Scholar] [CrossRef] [PubMed]
- Thingholm, L.B.; Rühlemann, M.C.; Koch, M.; Fuqua, B.; Laucke, G.; Boehm, R.; Bang, C.; Franzosa, E.A.; Hübenthal, M.; Frost, F.; et al. Obese Individuals with and without Type 2 Diabetes Show Different Gut Microbial Functional Capacity and Composition. Cell Host Microbe 2019, 26, 252–264.e10. [Google Scholar] [CrossRef] [PubMed]
- Gao, R.; Zhu, C.; Li, H.; Yin, M.; Pan, C.; Huang, L.; Kong, C.; Wang, X.; Zhang, Y.; Qu, S.; et al. Dysbiosis Signatures of Gut Microbiota Along the Sequence from Healthy, Young Patients to Those with Overweight and Obesity. Obesity 2018, 26, 351–361. [Google Scholar] [CrossRef] [PubMed]
- Armougom, F.; Henry, M.; Vialettes, B.; Raccah, D.; Raoult, D. Monitoring bacterial community of human gut microbiota reveals an increase in Lactobacillus in obese patients and Methanogens in anorexic patients. PLoS ONE 2009, 4, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Horie, M.; Miura, T.; Hirakata, S.; Hosoyama, A.; Sugino, S.; Umeno, A.; Murotomi, K.; Yoshida, Y.; Koike, T. Comparative analysis of the intestinal flora in type 2 diabetes and nondiabetic mice. Exp. Anim. 2017, 66, 405–416. [Google Scholar] [CrossRef] [Green Version]
- Larsen, N.; Vogensen, F.K.; Van Den Berg, F.W.J.; Nielsen, D.S.; Andreasen, A.S.; Pedersen, B.K.; Al-Soud, W.A.; Sørensen, S.J.; Hansen, L.H.; Jakobsen, M. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS ONE 2010, 5, e9085. [Google Scholar] [CrossRef]
- Sedighi, M.; Razavi, S.; Navab-Moghadam, F.; Khamseh, M.E.; Alaei-Shahmiri, F.; Mehrtash, A.; Amirmozafari, N. Comparison of gut microbiota in adult patients with type 2 diabetes and healthy individuals. Microb. Pathog. 2017, 111, 362–369. [Google Scholar] [CrossRef]
- Moghadam, F.; Sedighi, M.; Khamseh, M.E.; Alaei-Shahmiri, F.; Talebi, M.; Razavi, S.; Amirmozafari, N. The association of type II diabetes with gut microbiota composition. Microb. Pathog. 2017, 110, 630–636. [Google Scholar] [CrossRef]
- Ahmad, A.; Yang, W.; Chen, G.; Shafiq, M.; Javed, S.; Zaidi, S.S.A.; Shahid, R.; Liu, C.; Bokhari, H. Analysis of gut microbiota of obese individuals with type 2 diabetes and healthy individuals. PLoS ONE 2019, 14, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ejtahed, H.S.; Hoseini-Tavassol, Z.; Khatami, S.; Zangeneh, M.; Behrouzi, A.; Ahmadi, B.S.; Moshiri, A.; Hasani-Ranjbar, S.; Soroush, A.R.; Vaziri, F.; et al. Main gut bacterial composition differs between patients with type 1 and type 2 diabetes and non-diabetic adults. J. Diabetes Metab. Disord. 2020, 19, 265–271. [Google Scholar] [CrossRef] [PubMed]
- Takagi, T.; Naito, Y.; Kashiwagi, S.; Uchiyama, K.; Mizushima, K.; Kamada, K.; Ishikawa, T.; Inoue, R.; Okuda, K.; Tsujimoto, Y.; et al. Changes in the gut microbiota are associated with hypertension, hyperlipidemia, and type 2 diabetes mellitus in Japanese subjects. Nutrients 2020, 12, 2996. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Jiang, X.; Cao, M.; Ge, J.; Bao, Q.; Tang, L.; Chen, Y.; Li, L. Altered fecal microbiota correlates with liver biochemistry in nonobese patients with non-alcoholic fatty liver disease. Sci. Rep. 2016, 6, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Sun, G.; Wang, Z.; Wu, W.; Guo, H.; Peng, L.; Wu, L.; Guo, X.; Yang, Y. Characteristics of fecal microbiota in non-alcoholic fatty liver disease patients. Sci. China Life Sci. 2018, 61, 770–778. [Google Scholar] [CrossRef] [PubMed]
- Shen, F.; Zheng, R.; Sun, X.; Ding, W.; Wang, X.; Fan, J. Gut microbiota dysbiosis in patients with nonalcoholic fatty liver disease. Hepatobiliary Pancreat. Dis. Int. 2017, 16, 375–381. [Google Scholar] [CrossRef]
- Raman, R.M.; Ahmed, I.; Gillevet, P.M.; Probert, C.S.; Ratcliffe, N.M.; Smith, S.; Greenwood, R.; Sikaroodi, M.; Lam, V.; Crotty, P.; et al. Fecal microbiome and volatile organic compound metabolome in obese humans with nonalcoholic fatty liver disease. Clin. Gastroenterol. Hepatol. 2013, 11, 868–875.e3. [Google Scholar] [CrossRef]
- Michail, S.; Lin, M.; Frey, M.R.; Fanter, R.; Paliy, O.; Hilbush, B.; Reo, N.V. Altered gut microbial energy and metabolism in children with non-alcoholic fatty liver disease. FEMS Microbiol. Ecol. 2015, 91, 1–9. [Google Scholar] [CrossRef]
- Nistal, E.; de Miera, L.E.S.; Pomar, M.B.; Sánchez-Campos, S.; García-Mediavilla, M.V.; Álvarez-Cuenllas, B.; Linares, P.; Olcoz, J.L.; Arias-Loste, M.T.; García-Lobo, J.M.; et al. An altered fecal microbiota profile in patients with non-alcoholic fatty liver disease (NAFLD) associated with obesity. Rev. Española Enferm. Dig. 2019, 111, 275–282. [Google Scholar] [CrossRef]
- Loomba, R.; Seguritan, V.; Li, W.; Long, T.; Klitgord, N.; Bhatt, A.; Dulai, P.S.; Caussy, C.; Bettencourt, R.; Highlander, S.K.; et al. Gut microbiome based metagenomic signature for non-invasive detection of advanced fibrosis in human nonalcoholic fatty liver disease. Cell Metab. 2017, 25, 1054–1062. [Google Scholar] [CrossRef]
- Del Chierico, F.; Nobili, V.; Vernocchi, P.; Russo, A.; De Stefanis, C.; Gnani, D.; Furlanello, C.; Zandonà, A.; Paci, P.; Capuani, G.; et al. Gut microbiota profiling of pediatric nonalcoholic fatty liver disease and obese patients unveiled by an integrated meta-omics-based approach. Hepatology 2017, 65, 451–464. [Google Scholar] [CrossRef]
- Da Silva, H.E.; Teterina, A.; Comelli, E.M.; Taibi, A.; Arendt, B.M.; Fischer, S.E.; Lou, W.; Allard, J.P. Nonalcoholic fatty liver disease is associated with dysbiosis independent of body mass index and insulin resistance. Sci. Rep. 2018, 8, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Mouzaki, M.; Comelli, E.M.; Arendt, B.M.; Bonengel, J.; Fung, S.K.; Fischer, S.E.; Mcgilvray, I.D.; Allard, J.P. Intestinal microbiota in patients with nonalcoholic fatty liver disease. Hepatology 2013, 58, 120–127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, L.; Baker, S.S.; Gill, C.; Liu, W.; Alkhouri, R.; Baker, R.D.; Gill, S.R. Characterization of gut microbiomes in nonalcoholic steatohepatitis (NASH) patients: A connection between endogenous alcohol and NASH. Hepatology 2013, 57, 601–609. [Google Scholar] [CrossRef] [PubMed]
- Boursier, J.; Mueller, O.; Barret, M.; Machado, M.; Fizanne, L.; Arauo-Perez, F.; Guy, C.D.; Seed, P.C.; Rawls, J.F.; David, L.A.; et al. The severity of NAFLD is associated with gut dysbiosis and shift in the metabolic function of the gut microbiota. Hepatology 2016, 63, 764–775. [Google Scholar] [CrossRef] [Green Version]
- Qin, N.; Yang, F.; Li, A.; Prifti, E.; Chen, Y.; Shao, L.; Guo, J.; Le Chatelier, E.; Yao, J.; Wu, L.; et al. Alterations of the human gut microbiome in liver cirrhosis. Nature 2014, 513, 59–64. [Google Scholar] [CrossRef] [PubMed]
- Lim, M.Y.; You, H.J.; Yoon, H.S.; Kwon, B.; Lee, J.Y.; Lee, S.; Song, Y.M.; Lee, K.; Sung, J.; Ko, G. The effect of heritability and host genetics on the gut microbiota and metabolic syndrome. Gut 2017, 66, 1031–1038. [Google Scholar] [CrossRef] [PubMed]
- Arumugam, M.; Raes, J.; Pelletier, E.; Le Paslier, D.; Yamada, T.; Mende, D.R.; Fernandes, G.R.; Tap, J.; Bruls, T.; Batto, J.-M.; et al. Enterotypes of the Human Gut Microbiome. Nature 2011, 473, 174–180. [Google Scholar] [CrossRef]
- Singh, R.K.; Chang, H.-W.; Yan, D.; Lee, K.M.; Ucmak, D.; Wong, K.; Abrouk, M.; Farahnik, B.; Nakamura, M.; Zhu, T.H.; et al. Influence of Diet on the Gut Microbiome and Implications for Human Health. J. Transl. Med. 2017, 15, 73. [Google Scholar] [CrossRef] [Green Version]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M.A.; et al. Diet Rapidly and Reproducibly Alters the Human Gut Microbiome. Nature 2014, 505, 559–563. [Google Scholar] [CrossRef] [Green Version]
- De Filippis, F.; Pellegrini, N.; Vannini, L.; Jeffery, I.B.; La Storia, A.; Laghi, L.; Serrazanetti, D.I.; Di Cagno, R.; Ferrocino, I.; Lazzi, C.; et al. High-Level Adherence to a Mediterranean Diet Beneficially Impacts the Gut Microbiota and Associated Metabolome. Gut 2016, 65, 1812–1821. [Google Scholar] [CrossRef] [PubMed]
- Qian, L.; Gao, R.; Hong, L.; Pan, C.; Li, H.; Huang, J.; Qin, H. Association analysis of dietary habits with gut microbiota of a native Chinese community. Exp. Ther. Med. 2018, 16, 856–866. [Google Scholar] [CrossRef] [Green Version]
- Lim, M.Y.; Rho, M.; Song, Y.-M.; Lee, K.; Sung, J.; Ko, G. Stability of Gut Enterotypes in Korean Monozygotic Twins and Their Association with Biomarkers and Diet. Sci. Rep. 2014, 4, 7348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patra, J.K.K.; Das, G.; Paramithiotis, S.; Shin, H.-S. Kimchi and Other Widely Consumed Traditional Fermented Foods of Korea: A Review. Front. Microbiol. 2016, 7, 1493. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Woo, H.D.; Shin, A.; Kim, J. Dietary Patterns of Korean Adults and the Prevalence of Metabolic Syndrome: A Cross-Sectional Study. PLoS ONE 2014, 9, e111593. [Google Scholar] [CrossRef]
- Dimidi, E.; Cox, S.R.; Rossi, M.; Whelan, K. Fermented Foods: Definitions and Characteristics, Impact on the Gut Microbiota and Effects on Gastrointestinal Health and Disease. Nutrients 2019, 11, 1806. [Google Scholar] [CrossRef] [Green Version]
- Eshel, G.; Shepon, A.; Makov, T.; Milo, R. Land, irrigation water, greenhouse gas, and reactive nitrogen burdens of meat, eggs, and dairy production in the United States. Proc. Natl. Acad. Sci. USA 2014, 19, 11996–12001. [Google Scholar] [CrossRef] [Green Version]
- Bergman, Å.; Becher, G.; Blumberg, B.; Bjerregaard, P.; Bornman, R.; Brandt, I.; Casey, S.; Frouin, H.; Giudice, L.; Heindel, J.; et al. Manufacturing doubt about endocrine disrupter science—A rebuttal of industry-sponsored critical comments on the UNEP/WHO report State of the Science of Endocrine Disrupting Chemicals 2012. Regul. Toxicol. Pharm. 2015, 73, 1007–1017. [Google Scholar] [CrossRef]
- Claus, S.P.; Guillou, H.; Ellero-Simatos, S. The gut microbiota: A major player in the toxicity of environmental pollutants? NPJ Biofilms Microbiomes 2016, 2, 16003, Erratum in 2017, 22, 17001. [Google Scholar] [CrossRef]
- Sharma, S.; Tripathi, P. Gut Microbiome and type 2 diabetes: Where we are and where to go? J. Nutr. Biochem. 2019, 63, 101–108. [Google Scholar] [CrossRef]
- Aguilera, M.; Gálvez-Ontiveros, Y.; Rivas, A. Endobolome, a new concept for determining the influence of Microbiota Disrupting Chemicals (MDC) in relation to specific endocrine pathogenesis. Front. Microbiol. 2020, 11, 578007. [Google Scholar] [CrossRef] [PubMed]
- Gálvez-Ontiveros, Y.; Páez, S.; Monteagudo, C.; Rivas, A. Endocrine Disruptors in Food: Impact on Gut Microbiota and Metabolic Diseases. Nutrients 2020, 12, 1158. [Google Scholar] [CrossRef] [PubMed]
- Darbre, P. Endocrine disruptors and obesity. Curr. Obes. Rep. 2017, 6, 18–27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vanhaecke, L.; Derycke, L.; Le Curieux, F.; Lust, S.; Marzin, D.; Verstraete, W.; Bracke, M. The microbial PhIP metabolite 7-hydroxy-5-methyl-3-phenyl-6,7,8,9-tetrahydropyrido [3′,2′:4,5] imidazo [1,2-a]pyrimidin-5-ium chloride (PhIP-M1) induces DNA damage, apoptosis and cell cycle arrest towards Caco-2 cells. Toxicol. Lett. 2008, 178, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Carmody, R.N.; Turnbaugh, P.J. Host-microbial interactions in the metabolism of therapeutic and diet-derived xenobiotics. J. Clin. Investig. 2014, 124, 4173–4181. [Google Scholar] [CrossRef]
- Kassie, F.; Laky, B.; Nobis, E.; Kundi, M.; Knasmüller, S. Genotoxic effects of methyl isothiocyanate. Mutat. Res. 2001, 490, 1–9. [Google Scholar] [CrossRef]
- Zhang, X.; Gao, B.; Creamer, A.E.; Cao, C.; Li, Y. Adsorption of VOCs onto engineered carbon materials: A review. J. Hazard. Mater. 2017, 338, 102–123. [Google Scholar] [CrossRef]
- Abdelsalam, N.A.; Ramadan, A.T.; El Rakaiby, M.T.; Aziz, R.K. Toxicomicrobiomics: The Human Microbiome vs. Pharmaceutical, Dietary, and Environmental Xenobiotics. Front. Pharm. 2020, 11, 390. [Google Scholar] [CrossRef]
- Hayashi, H.; Sakamoto, M.; Benno, Y. Phylogenetic Analysis of the Human Gut Microbiota Using 16S RDNA Clone Libraries and Strictly Anaerobic Culture-Based Methods. Microbiol. Immunol. 2002, 46, 535–548. [Google Scholar] [CrossRef]
- Martínez, N.; Hidalgo-Cantabrana, C.; Delgado, S.; Margolles, A.; Sánchez, B. Filling the Gap between Collection, Transport and Storage of the Human Gut Microbiota. Sci. Rep. 2019, 9, 88. [Google Scholar] [CrossRef] [Green Version]
- Stewart, E.J. Growing unculturable bacteria. J. Bacteriol. 2012, 194, 4151–4160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lagier, J.-C.; Dubourg, G.; Million, M.; Cadoret, F.; Bilen, M.; Fenollar, F.; Levasseur, A.; Rolain, J.-M.; Fournier, P.-E.; Raoult, D. Culturing the Human Microbiota and Culturomics. Nat. Rev. Microbiol. 2018, 16, 540–550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- López-Moreno, A.; Torres-Sánchez, A.; Acuña, I.; Suárez, A.; Aguilera, M. Representative Bacillus sp. AM1 from gut microbiota harbor versatile molecular pathways for Bisphenol A biodegradation. Int. J. Mol. Sci. 2021, 22, 4952. [Google Scholar] [CrossRef]
- Browne, H.P.; Forster, S.C.; Anonye, B.O.; Kumar, N.; Neville, B.A.; Stares, M.D.; Goulding, D.; Lawley, T.D. Culturing of ‘unculturable’ human microbiota reveals novel taxa and extensive sporulation. Nature 2020, 533, 543–546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, Y.; Hou, F.; Pan, Z.; Huang, Z.; Han, N.; Bin, L.; Deng, H.; Li, Z.; Ding, L.; Gao, H.; et al. Optimization of culturomics strategy in human fecal samples. Front. Microbiol. 2019, 10, 2891. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gotoh, A.; Nara, M.; Sugiyama, Y.; Sakanaka, M.; Yachi, H.; Kitakata, A.; Nakagawa, A.; Minami, H.; Okuda, S.; Katoh, T.; et al. Use of Gifu Anaerobic Medium for culturing 32 dominant species of human gut microbes and its evaluation based on short-chain fatty acids fermentation profiles. Biosci. Biotechnol. Biochem. 2017, 81, 2009–2017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lagier, J.-C.; El Karkouri, K.; Mishra, A.K.; Robert, C.; Raoult, D.; Fournier, P.-E. Non contiguous-finished genome sequence and description of Enterobacter massiliensis sp. nov. Stand. Genom. Sci. 2013, 7, 399–412. [Google Scholar] [CrossRef] [Green Version]
- Bailey, M.T.; Coe, C.L. Endometriosis is associated with an altered profile of intestinal microflora in female rhesus monkeys. Hum. Reprod. 2002, 17, 1704–1708. [Google Scholar] [CrossRef] [Green Version]
- Lei, M.; Menon, R.; Manteiga, S.; Alden, N.; Hunt, C.; Alaniz, R.C.; Lee, K.; Jayaraman, A. Environmental chemical diethylhexyl phthalate alters intestinal microbiota community structure and metabolite profile in mice. Msystems 2019, 4, 00724-19. [Google Scholar] [CrossRef] [Green Version]
- Williams, N.T. Probiotics. Am. J. Health Sys. Pharm. 2010, 67, 449–458. [Google Scholar] [CrossRef]
- Farnworth, E.R. The evidence to support health claims for probiotics. J. Nutr. 2008, 138, 1250–1254. [Google Scholar] [CrossRef] [PubMed]
- Ouwehand, A.C. A Review of Dose-Responses of Probiotics in Human Studies. Benef. Microbes 2017, 8, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Ouwehand, A.C.; Invernici, M.M.; Furlaneto, F.A.C.; Messora, M.R. Effectiveness of Multistrain Versus Single-Strain Probiotics: Current Status and Recommendations for the Future. J. Clin. Gastroenterol. 2018, 52 (Suppl. 1), S35–S40. [Google Scholar] [CrossRef]
- López-Moreno, A.; Aguilera, M. Vaginal probiotics for reproductive health and related dysbiosis: Sistematic review and meta-analysis. J. Clin. Med. 2021, 1461. [Google Scholar] [CrossRef]
- López-Moreno, A.; Suárez, A.; Avanzi, C.; Monteoliva-Sánchez, M.; Aguilera, M. Probiotic Strains and Intervention Total Doses for Modulating Obesity-Related Microbiota Dysbiosis: A Systematic Review and Meta-Analysis. Nutrients 2020, 12, 1921. [Google Scholar] [CrossRef]
- Rijkers, G.T.; Bengmark, S.; Enck, P.; Haller, D.; Herz, U.; Kalliomaki, M.; Kudo, S.; Lenoir-Wijnkoop, I.; Mercenier, A.; Myllyluoma, E.; et al. Guidance for substantiating the evidence for beneficial effects of probiotics: Current status and recommendations for future research. J. Nutr. 2010, 140, 671S–676S. [Google Scholar] [CrossRef] [Green Version]
- Miyoshi, J.; Rao, M.C.; Chang, E.B. Navigating the human gut microbiome: Pathway to success from lessons learned. Gastroenterology 2020, 159, 2019–2024. [Google Scholar] [CrossRef]
- Amrane, S.; Hocquart, M.; Afouda, P.; Kuete, E.; Pham, T.P.T.; Dione, N.; Ngom, I.I.; Valles, C.; Bachar, D.; Raoult, D.; et al. Metagenomic and culturomic analysis of gut microbiota dysbiosis during Clostridium difficile infection. Sci. Rep. 2019, 9, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Dhakan, D.B.; Maji, A.; Sharma, A.K.; Saxena, R.; Pulikkan, J.; Grace, T.; Gomez, A.; Scaria, J.; Amato, K.R.; Sharma, V.K. The unique composition of Indian gut microbiome, gene catalogue, and associated fecal metabolome deciphered using multi-omics approaches. GigaScience 2019, 8, 1–20. [Google Scholar] [CrossRef]
- Egidius, H.M.; Veidal, S.S.; Feigh, M.; Hallenborg, P.; Puglia, M.; Pers, T.H.; Vrang, N.; Jelsing, J.; Kornum, B.R.; Blagoev, B.; et al. Multi-omics characterization of a diet-induced obese model of non-alcoholic steatohepatitis. Sci. Rep. 2020, 10, 1148. [Google Scholar] [CrossRef]
- Graw, S.; Chappell, K.; Washam, L.C.; Gies, A.; Bird, J.; Robeson, S.M.; Byrum, S.D. Multi-omics data integration considerations and study design for biological systems and disease. Mol. Omics 2021, 17, 170–185. [Google Scholar] [CrossRef]
- Jiménez-Pranteda, M.L.; Pérez-Davó, A.; Monteoliva-Sánchez, M.; Ramos-Cormenzana, A.; Aguilera, M. Food Omics Validation: Towards Understanding Key Features for Gut Microbiota, Probiotics and Human Health. Food Anal. Methods 2015, 8, 272–289. [Google Scholar] [CrossRef]
- Valles-Colomer, M.; Darzi, Y.; Vieira-Silva, S.; Falony, G.; Raes, J.; Joossens, M. Meta-omics in inflammatory bowel disease research: Applications, challenges, and guidelines. J. Crohns. Colitis 2016, 10, 735–746. [Google Scholar] [CrossRef] [Green Version]
- Mallick, H.; Bucci, V.; An, L. Experimental design and quantitative analysis of microbial community multiomics. Genome Biol. 2017, 18, 1–16. [Google Scholar] [CrossRef]
- Knight, R.; Vrbanac, A.; Taylor, B.C.; Aksenov, A.; Callewaert, C.; Debelius, J.; Gonzalez, A.; Kosciolek, T.; McCall, L.I.; McDonald, D.; et al. Best practices for analysing microbiomes. Nat. Rev. Microbiol. 2018, 16, 410–422. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Li, L.; Butcher, J.; Stintzi, A.; Figeys, D. Advancing functional and translational microbiome research using meta-omics approaches. Microbiome 2019, 7, 1–12. [Google Scholar] [CrossRef]
- The European Parliament and the Council of the European Union. Regulation (EC) No 258/97 of the European Parliament and of the Council of 27 January 1997 concerning novel foods and novel food ingredients. Off. J. Eur. Union 1997, 43, 0001–0006. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/?uri=celex%3A31997R0258 (accessed on 10 February 2021).
- The European Parliament and the Council of the European Union. Regulation (EC) No 1924/2006 of the European Parliament and the of the Council of 20 December 2006 on nutrition and health claims made on foods. Off. J. Eur. Union 2006, 404, 9–25. Available online: https://eur-lex.europa.eu/legal-content/en/ALL/?uri=CELEX%3A32006R1924 (accessed on 10 February 2021).
- Cordaillat-Simmons, M.; Rouanet, A.; Pot, B. Live biotherapeutic products: The importance of a defined regulatory framework. Exp. Mol. Med. 2020, 52, 1397–1406. [Google Scholar] [CrossRef]
- Ricci, A.; Allende, A.; Bolton, D.; Chemaly, M.; Davies, R.; Girones, R.; Herman, L.; Koutsoumanis, K.; Lindqvist, R.; Nørrung, B.; et al. Scientific Opinion on the update of the list of QPS-recommended biological agents intentionally added to food or feed as notified to EFSA. EFSA J. 2017, 15, 3. [Google Scholar] [CrossRef] [Green Version]
- United States Food and Drug Administration. Federal Food, Drug, and Cosmetic Act; United States Food and Drug Administration (FDA), 2018. Available online: http://www.fda.gov/regulatoryinformation/legislation/federalfooddrugandcosmeticacfdcact/default.htm/ (accessed on 10 March 2021).
- Degnan, F.H. Clinical studies involving probiotics: When FDA’s investigational new drug rubric applies-and when it may not. Gut Microbes 2012, 3, 485–489. [Google Scholar] [CrossRef] [Green Version]
- Yang, M.S. Regulatory aspects of nutraceuticals: Chinese perspective. In Nutraceuticals, 1st ed.; Grumezescu, A., Ed.; Elsevier: New York, NY, USA, 2016; pp. 947–957. [Google Scholar]
- de Simone, C. The Unregulated Probiotic Market. Clin. Gastroenterol. Hepatol. 2019, 17, 809–817. [Google Scholar] [CrossRef] [Green Version]
- Amagase, H. Current marketplace for probiotics: A Japanese perspective. Clin. Infect. Dis. 2008, 46, 73–75. [Google Scholar] [CrossRef] [Green Version]
- Nagata, J.; Yamada, K. Foods with Health Claims in Japan. Food Sci. Technol. Res. 2008, 14, 519–524. [Google Scholar] [CrossRef]
- Arora, M.; Baldi, A. Comparative study of regulatory framework for probiotics: Current status and future recommendations. Appl. Clin. Res. Clin. Trials Regul. Aff. 2017, 4, 140–156. [Google Scholar] [CrossRef]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. Expert consensus document: The international scientific association for probiotics and prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef] [Green Version]
- Lin, T.L.; Shu, C.C.; Lai, W.F.; Tzeng, C.M.; Lai, H.C.; Lu, C.C. Investiture of next generation probiotics on amelioration of diseases–Strains do matter. Med. Microecol. 2019, 2, 100002. [Google Scholar] [CrossRef]
- Saarela, M.H. Safety aspects of next generation probiotics. Curr. Opin. Food Sci. 2019, 30, 8–13. [Google Scholar] [CrossRef]
- Morovic, W.; Roper, J.M.; Smith, A.B.; Mukerji, P.; Stahl, B.; Rae, J.C.; Ouwehand, A.C. Safety evaluation of HOWARU® Restore (Lactobacillus acidophilus NCFM, Lactobacillus paracasei Lpc-37, Bifidobacterium animalis subsp. lactis Bl-04 and B. lactis Bi-07) for antibiotic resistance, genomic risk factors, and acute toxicity. Food Chem. Toxicol. 2017, 110, 316–324. [Google Scholar] [CrossRef]
- Li, B.; Zhan, M.; Evivie, S.E.; Jin, D.; Zhao, L.; Chowdhury, S.; Sarker, S.K.; Huo, G.; Liu, F. Evaluating the safety of potential probiotic Enterococcus durans KLDS6.0930 using whole genome sequencing and oral toxicity study. Front. Microbiol. 2018, 9, 1–15. [Google Scholar] [CrossRef] [Green Version]
- The European Parliament; The Council of the European Union. Directive 2000/13/EC on the Approximation of the Laws of the Member States Relating to the Labelling, Presentation and Advertising of Foodstuffs. Off. J. Eur. Union 2000, 109, 29–42. Available online: https://eur-lex.europa.eu/legal-content/EN/ALL/?uri=celex%3A32000L0013 (accessed on 10 February 2021).
- The European Parliament; The Council of the European Union. Regulation (EU) No 1169/2011 of the European Parliament and of the Council of 25 October 2011 on the Provision of Food Information to Consumers, Amending Regulations (EC) No 1924/2006 and (EC) No 1925/2006 of the European Parliament and of the Council, and Repealing Commission Directive 87/250/EEC, Council Directive 90/496/EEC, Commission Directive 1999/10/EC, Directive 2000/13/EC of the European Parliament and of the Council, Commission Directives 2002/67/EC and 2008/5/EC and Commission Regulation (EC) No 608/2004 Off. J. Eur. Union 2011, 27, 18–63. Available online: https://eur-lex.europa.eu/legal-content/EN/ALL/?uri=CELEX%3A32011R1169 (accessed on 10 February 2021).
- Hoffmann, D.E. Health claim regulation of probiotics in the USA and the EU: Is there a middle way? Benef. Microbes 2013, 4, 109–115. [Google Scholar] [CrossRef]
- International Probiotics Association. Best Practices Guidelines for Probiotics. Int. Probiotics Assoc. 2017, 1–4. Available online: http://internationalprobiotics.org/wp-content/uploads/CRN-IPA-Best-Practices-Guidelines-for-Probiotics-final.pdf (accessed on 10 February 2021).
- Reid, G.; Kort, R.; Alvarez, S.; Bourdet-Sicard, R.; Benoit, V.; Cunningham, M.; Saulnier, D.M.; Van Hylckama, V.J.E.T.; Verstraelen, H.; Sybesma, W. Expanding the reach of probiotics through social enterprises. Benef. Microbes 2018, 9, 707–715. [Google Scholar] [CrossRef] [Green Version]
- International Scientific Association for Probiotics and Prebiotics. Probiotic Product Labels. Int. Sci. Assoc. Probiotics Prebiotics 2017. Available online: https://isappscience.org/for-consumers/probiotic-product-labels/ (accessed on 10 February 2021).
- Wang, J.; Pourang, A.; Burrall, B. Open access medical journals: Benefits and challenges. Clin. Derm. 2019, 37, 52–55. [Google Scholar] [CrossRef]


| NGP Microbial Strains, Target Species, Study Reference | Study Design | Dietary Aspects | Clinical Effects and Functionality |
|---|---|---|---|
| Akkermansia muciniphila Muc [CIP 107961]—Human [41] [ClinicalTrials.gov Identifier: NCT02637115] | ICT: randomized, double-blind, placebo-controlled pilot study Live probiotics 1010/day vs. pasteurized probiotics 1010/day vs. placebo in patients with metabolic syndrome | Normal dietary intake and physical activity during the study period | ↑ Insulin sensitivity, ↓ insulinemia and ↓plasma total cholesterol |
| Akkermansia muciniphila WST01—Human [42] [ClinicalTrials.gov Identifier: NCT04797442] | ICT: randomized, double-blind, placebo-controlled, multicenter clinical trial Probiotics vs. placebo in overweight or obese patients with type 2 diabetes | Intervention added onto lifestyle | Results will be available in June 2022 |
| Christensenella minuta Xla1—Human [43] [ClinicalTrials.gov Identifier: NCT04663139] | ICT: randomized, partially placebo-controlled double-blind Probiotics vs. placebo in healthy volunteers, overweight, and obese adults | Agreement to keep food, drink, physical activities, and alcohol consumption habits unchanged throughout the study | Results will be available in October 2021 |
| Eubacterium hallii—Human [44] [ClinicalTrials.gov Identifier: NCT04529473] | ICT:double-blind, randomized, placebo-controlled Probiotics vs. placebo | Maintenance of dietary habits and physical activity levels throughout the study period | Results will be available on January 2022 |
| Hafnia alvei HA4597—Human [45] [ClinicalTrials.gov Identifier: NCT03657186] | ICT: multicenter, randomized, double-blind placebo-controlled study. Probiotics vs. placebo on weight reduction in overweight subjects | −20% hypocaloric diet and maintainance of the usual physical activity | ↑ Weight loss in overweight subjects, ↑ feeling of fullness, ↑ loss of hip circumference, ↓ fasting glycemia |
| Lactococcus lactis NRRL-B50571—Human [46] [ClinicalTrials.gov Identifier: NCT02670811] | ICT: double-blind randomized controlled Probiotics vs. placebo on prehypertensive subjects | Participants were asked not to change their diet or lifestyle during the intervention | ↓ Systolic and diastolic blood pressure, ↓ Triglyceride, total cholesterol, and low-density lipoprotein |
| Escherichia coli Nissle 1917—Human [47] [ClinicalTrials.gov Identifier: NCT02144948] | ICT: single group assignment. Patients with type 2 diabetes | - | Results not yet available or posted on ClinicalTrials.gov November 2021 |
| Akkermansia muciniphila—Muc [CIP 107961]—Mice [48,49] | PCS: probiotics vs. control. Obesity | High-fat diet/standard diet | ↓ Fat-mass gain, ↑ insulin sensitivity, restore gut barrier function by acting on TLR2, ↑ mucus later thickness; similar effects by a purified membrane protein alone (Amuc_1100) |
| Clostridium butyricum CGMCC0313.1—Mice [50] | PCS: probiotics vs. control. Obesity | High-fat diet/standard diet | ↓ Lipid accumulation in liver and serum, ↓ insulin levels, ↑ glucose tolerance, ↑ insulin sensitivity, ↓ TNF-α and ↑ IL-10 and IL-22 in colon |
| Faecalibacterium prausnitzii VPI C13-20-A—Mice [51] | PCS: probiotics vs. control. Obesity | High-fat diet/standard diet | ↑ Hepatic health, ↓ adipose tissue inflammation |
| Bacteroides uniformis CECT 7771– Mice [52] | PCS: probiotics vs. control. Obesity | High-fat diet/standard diet | ↓ Weight gain; ↓ dietary fat absorption; ↓ liver steatosis; ↓ serum cholesterol, triglyceride, glucose, insulin and leptin; ↑ glucose tolerance; ↑ TNF-α by DCs after LPS stimulation;↑ phagocytosis |
| Parabacteroides goldsteinii JCM 13446—Mice [53] | PCS: probiotics vs. control. Obesity | High-fat diet/standard diet | ↓ Obesity by ↑ adipose tissue thermogenesis, ↑ intestinal integrity ↓ inflammation, ↑ insulin sensitivity |
| Christensenella minuta—Mice [54] | PCS: probiotics vs. control. Obesity | High-fat diet/standard diet | ↓ Weight gain, ↓ adiposity. Highly heritable in a lean host phenotype |
| Eubacterium hallii DSM 17630—Mice [55] | PCS: probiotics vs. control. Diabetes | High-fat diet/standard diet | ↑ Energy metabolism and ↑ insulin sensitivity through glycerol conversion 3hydroxypropionaldehyde |
| Hafnia alvei HA4597—Mice [56] | PCS: probiotics vs. control. Obesity | High-fat diet/standard diet | ↑ Beneficial anti-obesity and metabolic effects, ↓ food intake, ↓ body weight and ↓ fat mass gain |
| Lactococcus lactis (GMM) LL-pCYT: HSP65-6P277 and LL-pHJ—Mice [57] | PCS: probiotics vs. control. Obesity | High-fat diet/standard diet | ↓ Antigen-specific of cellular immunity |
| Escherichia coli Nissle 1917 (EcN-GMM)– Mice [58] | PCS: probiotics vs. control. Obesity | High-fat diet/standard diet | Modulation of the neuropeptide expression of energy intake and expenditure in the hypothalamus |
| Reference | Subjects and Disease | Dietary Aspects | Sample Size and Clinical Traits | Detection Technique | Microbial Taxa Modifications |
|---|---|---|---|---|---|
| Zhong et al. [66] | Human Obesity | NA | N = 382; MHNO n = 191; MUNO n = 61; MHO n = 66; MUO n = 64 | MiSeq platform (Illumina) V3–V4 region of the 16S rRNA gene | ↑ Lachnospiraceae, Bacteroidaceae, Methanobacteriaceae and Pasteurellaceae in MHNO and MUNO |
| Jonduo et al. [67] | Human Obesity | Participant’s predominantly plant-based diet: vegetables (e.g., sweet potato, cassava, plantain, and beans) | n = 18; OB n = 9; Non-OB n = 9 | 454 GS FLX platform or 454 GS JUNIOR system (Roche) V1-V2 region of the 16S rRNA gene | ↑ Prevotella in almost all individuals |
| Thingholm et al. [68] | Human Obesity | NA | n = 1280; LH n = 633; OBH n = 494; OBT2D n = 153 | MiSeq platform (Illumina) V1-V2 region of 16S rRNA gene | ↓ Akkermansia, Faecalibacterium, Oscillibacter, and Alistipes in obese individuals ↓ Faecalibacterium prausnitzii in obese individuals |
| Schwiertz et al. [65] | Human Obesity | Western diet | n= 98; HC n = 30; OW n = 35; OB n = 33 | qPCR | ↑ Bacteroides in overweight vs. HC ↓ Ruminococcus flavefaciens in overweight and obese ↓ Bifidobacterium and Clostridium leptum in obese ↓ Methanobrevibacter in overweight and obese |
| Gao et al. [69] | Human Obesity | NA | n = 192; HC n = 25; OW n = 22; OB n = 145 | MiSeq platform (Illumina) V4 region of the 16S rRNA gene | ↑ Lachnoclostridium, Fusobacterium, Escherichia-Shigella, Klebsiella, Bacillus, and Pseudomonas in OW and OB ↑ Clostridia, Faecalibacterium, Ruminococcus, Bifidobacterium, and Lachnospiraceae_UCG_008 in HC |
| Armougom et al. [70] | Human Obesity Anorexia nervosa | NA | n= 49; HC n = 20; OB n = 20; AN n = 9 | qPCR | ↑ Lactobacillus in OB |
| Horie et al. [71] | Mice Type 2 diabetes | NA | 5-week-old TSNO mice n = 5; 5-week-old TSOD mice n = 5; 12-week-old TSNO mice n = 5; 12-week-old TSOD mice n = 5 | qPCR | ↑ Lactobacillus in TSOD vs. TSNO ↑ Bacteroidales and Lachnospiraceae in TSNO vs. TSOD ↑ Turicibacter and SMB53 in TSOD |
| Larsen et al. [72] | Human Type 2 diabetes | NA | n = 36; HC n = 18; T2D n = 18 | MiSeq platform (Illumina) V4 region of the 16S rRNA gene | ↑ Firmicutes in HC ↑ Bacteroidetes and Betaproteobacteria in T2D ↓ Clostridia in T2D |
| Sedighi et al. [73] | Human Type 2 diabetes | NA | n = 36; HC n = 18; T2D n = 18 | qPCR | ↑ Lactobacillus in T2D ↑ Bifidobacterium in HC ↑ Fusobacterium in T2D |
| Moghadam et al. [74] | Human Tipe 2 diabetes | NA | n = 36; HC n = 18; T2D n = 18 | qPCR | ↑ Faecalibacterium prausnitzii in HC |
| Ahmad et al. [75] | Human Type 2 diabetes Obesity | Eastern dietary habits (high carbohydrate and fat intake, low fiber intake) | n = 60; HC n = 20; Obese-T2D n = 40 | MiSeq platform (Illumina) V3–V4 region of the 16S rRNA gene | ↑ Firmicutes in Obese-T2D ↑ Clostridia, Negativicutes, Coriobacteria, Acidobacteria, Deferribacteres, and Gemmatimonadetes in obese-T2D ↑ Verrucomicrobia, Bacteroidetes, Proteobacteria, and Elusimicrobia in HC ↑ Prevotella P4_76, Clostridiales, Porphyromonadaceae bacterium DJF B175, Candidatus Alistipes marseilloanorexic AP11, Bacillus sporothermodurans, Staphylococcus SV3, and Iamia in obese-T2D |
| Ejtahed et al. [76] | Human Type 2 diabetes Type 1 diabetes | NA | n = 110; HC n = 40; T2D n = 49; T1D n = 21 | qPCR | ↑ Escherichia, Prevotella, and Lactobacillus in T1D and T2D ↑ Bifidobacterium, Roseburia, and Bacteroides in HC ↓ Faecalibacterium in T1D vs. HC and T2D |
| Takagi et al. [77] | Human Type 2 diabetes Hypertension Hyperlipidemia | NA | n = 239; HC n = 54; HT n = 97; HL n = 96; T2D n = 162 | MiSeq platform (Illumina) V3–V4 region of the 16S rRNA gene | ↑ Actinobacteria in HT, HL, T2D, RISK2, and RISK3 ↓ Bacteroidetes in HT, HL, T2D and RISK3 ↑ Bifidobacterium in HL, T2D, RISK1 and RISK2 ↑ Collinsella in HT, HL, T2D, RISK2 and RISK3 ↑ Escherichia in RISK 3 ↓ Alistipes in HL |
| Wang et al. [78] | Human Non-alcoholic fatty liver disease | Omnivorous Chinese diet | n = 126; HC n = 83; NAFLD n = 43 | 454 Life Sciences Genome Sequencer FLX system (Roche) V3 region of the 16S rRNA gene | ↓ Firmicutes ↑Bacteroidetes in NAFLD ↑ Bacteroidia ↓ Clostridia in NAFLD ↓ Coprococcus, Pseudobutyrivibrio, Moryella, Roseburia, Anaerotruncus, Ruminococcus, Anaerosporobacter, andLactobacillus in NAFLD |
| Li et al. [79] | Human Non-alcoholic fatty liver disease | No dietary restrictions imposed | n = 67; HC n = 37; NAFLD n = 30 | MiSeq platform (Illumina) V4 region of the16S rRNA gene | ↑ Lactobacillaceae, Peptostreptococcaceae, Veillonellaceae, EtOH8, Coprobacillaceae, and Erysipelotrichaceae in NAFLD ↑ Porphyromonas and Succinivibrio in NAFLD ↓ Odoribacter and Proteus in NAFLD |
| Shen et al. [80] | Human Non-alcoholic fatty liver disease | NA | n = 47; HC n = 22; NAFLD n = 25 | 454 GS-FLX platform (Roche) V3-V5 region of the 16S rRNA gene | ↑ Proteobacteria, Fusobacteria, Lachnospiraceae_Incertae_Sedis and Blautia in NAFLD ↑ Bacteroidetes and Prevotella in HC ↑ Escherichia_Shigella, Clostridium_XVIII, and Staphylococcus in NAFLD |
| Raman et al. [81] | Human Non-alcoholic fatty liver disease | No dietary restrictions imposed | n = 60; HC n = 30; NAFLD n = 30 | qPCR | ↑ Lactobacillus, Roseburia, Dorea, and Robinsoniella in NAFLD ↓Oscillibacterin NAFLD |
| Michail et al. [82] | Human Non-alcoholic fatty liver disease Obesity | No dietary restrictions imposed | n = 50; HC n = 26; NAFLD n = 13; Obese non-NAFLD n = 11 | qPCR | ↑ Gammaproteobacteria, Prevotella, and Epsilonproteobacteria in NAFLD ↓ Clostridia ↑ Alphaproteobacteria in obese non-NAFLD |
| Nistal et al. [83] | Human Non-alcoholic fatty liver disease Obesity | NA | n = 73; HC n = 20; Obese-NAFLD n = 36; Obese non-NAFLD n = 17 | MiSeq platform (Illumina) V3–V4 region of the 16S rRNA gene | ↑ Bacilli in obese-NAFLD ↓ Betaproteobacteria in obese-NAFLD vs. obese non-NAFLD ↓ Oscillospira, Akkermansia, and Eubacterium in obese-NAFLD and obese non-NAFLD vs. HC ↑ Megasphaera, Lactobacillus, Acidominococcus in obese-NAFLD, and obese non-NAFLD vs. HC ↓ Blautia, Alkaliphilus, and Flavobacterium in obese-NAFLD ↑ Staphylococcus in obese-NAFLD |
| Loomba et al. [84] | Human Non-alcoholic fatty liver disease Fibrosis | NA | n= 86; NAFLD n = 72; Fibrosis n = 14 | qPCR | ↑ Firmicutes in NAFLD, ↑ Proteobacteria in fibrosis ↑ Eubacterium rectale and Bacteroides vulgatus in NAFLD ↑ Bacteroides vulgatus and Escherichia coli in fibrosis ↓ Ruminococcus obeum, and Eubacterium rectale in fibrosis |
| Del Chierico et al. [85] | Human Non-alcoholic fatty liver disease Non-alcoholic steatohepatitis Obesity | NA | n= 115; HC n = 54, OB n = 8; NAFLD n = 27; NASH n = 26 | 454- Junior Genome Sequencer FLX system (Roche) V1-V3 region of the 16S rRNA gene | ↑ Bradyrhizobium, Anaerococcus, Peptoniphilus, Propionibacterium acnes, Dorea, and Ruminococcus ↓ Oscillospira and Rikenellaceae in NAFLD ↑ Ruminococcus, Dorea, and Blautia in NASH |
| Da Silva et al. [86] | Human Non-alcoholic steatohepatitis Simple steatosis | 7-day food record | n = 67; HC n = 28; SS n = 15: NASH n = 24 | MiSeq platform (Illumina) | ↓ Ruminococcus, Faecalibacteriumprausnitzii, and Coprococcus in NASH and SS vs. HC |
| Mouzaki et al. [87] | Human Non-alcoholic steatohepatitis Simple steatosis | HC patients were consuming more calories per kg compared to patients with NASH | n = 50; HC n = 17; SS n = 11; NASH n = 22 | qPCR | ↓ Bacteroidetes in NASH vs. SS and HC ↑ Clostridium coccoides in NASH vs. SS |
| Zhu et al. [88] | Human Non-alcoholic steatohepatitis Obesity | NA | n= 63; HC n = 16; OB n = 25; NASH n = 22 | qPCR | ↑ Bacteroides ↓ Firmicutes in NASH and OB ↓ Blautia and Faecalibacterium in NASH and OB |
| Boursier et al. [89] | Human Non-alcoholic steatohepatitis Fibrosis | NA | n = 57; Non-NASH n = 20 NASH n = 10; Fibrosis ≥ 2 n = 27 | Illumina V4 region of 16S rRNA gene | ↑ Bacteroides ↓Prevotella in NASH ↑ Bacteroides and Ruminococcus in fibrosis ≥ 2 ↓ Prevotella in fibrosis ≥ 2 |
| Qin et al. [90] | Human Cirrhosis | NA | n= 179; HC n = 83; Cirrhosis n = 96 | qPCR | ↑ Streptococcus, Veillonella, Clostridium and Prevotella in cirrhosis ↑ Eubacterium and Alistipes in HC ↓ Bacteroides in cirrhosis |
| Lim et al. [91] | Human Methabolic syndrome | NA | n = 655; Monozygotic twins n = 306; Dizygotic twins n = 74; Siblings n = 275 | MiSeq platform (Illumina) V4 region of the 16S rRNA gene | ↑ Lactobacillus, Sutterella and Methanobrevibacter in MetS ↓ Parabacteroides, Bifidobacterium, Odoribacter, Akkermansia and Christensenella in MetS |
| Reference/Sample | Culture Media | Culture Media Modifications | Selected Favored Cultured Microorganisms | Outcome and Observations: New Species Cultured: Potential NGP |
|---|---|---|---|---|
| Browne et al. [118] Human | YCFA | Glucose (0.2%), maltose (0.2%), and cellobiose (0.2%) | Aero-intolerant genus and species | 68 new isolated species: 16S RNA similarity 86–97% Anaerotruncus colihominis Blautia luti; B. hydrogenotrophica Clostridium boltae; C. celerecrescens; C. celerescens; C. clostridioforme; C. cocleatum; C. disporicum; C. ghonii; C. hathewayi; C. innocuum; C. lituseburense; C. methylpentosum; C. nexile; C. oroticum; C. saccharogumia; C. saccharolyticum; C. thermocellum; C. xylanolyticum Coprococcus eutactus Oscillibacter valericigenes Roseburia faecis; R. inulinivorans Ruminococcus albus; R.bromii; R. flavefaciens; R. gnavus; R.obeum; R. torques |
| YCFA | Pre-treatment with ethanol 70% (v/v), glucose (0.2%), maltose (0.2%), cellobiose (0.2%), sodium taurocholate (0.1%). Spore-forming gut aero-intolerant bacteria | Alistipes finegoldii Anaerotruncus colihominis Blautia hydrogenotrophica; B. obeum; B. wexlerae Clostridum baratti; C. bartlettii; C. clostridioforme; C. disporicum; C. hathewayi; C.innocuum; C. paraputrificum; C.perfringens Coprococcus comes; C. eutactus Prevotella copri Roseburia hominis; R. intestinalis; R. inulinvorans; Ruminococcus bromii; R. gnavus; R. obeum; R. torques | ||
| Chang et al. [119] Human | YCFA | Pre-incubation in blood culture bottles supplemented with 10% sheep blood and 10% rumen | Aero-intolerant bacteria Alistipes shahii; A. onderdonkii, Clostridium bifermentans, C. innocuum, C. hiranonis, C. butiricum, C. hathewayi, C. bolteae, C. sporogenes, Odoribacter splanchnicus | 22% of species isolated increase: 16S RNA similarity 93–97% 3 new species isolated: Longicatena caemuris Bacillus alcalophilus Pseudogracilibacillus auburnensis |
| Gotoh et al. [120] Microbial bank | GAM | NA | Aero-intolerant bacteria 72% of species of the top 56 species listed in the “human gut microbial gene catalogue” cultured in GAM | Isolated species in GAM: Anaerotruncus colihominis, Blautia hansenii, Clostridium nexile, C. asparagiforme, C. scindens, Coprococcus comes Roseburia intestinalis Ruminococcus torques, R. lactaris, R. obeum, R. gnavus. |
| Lagier et al. [121] 16-years-old male | BHI | Preincubation of the stool with lytic E. coli T1 and T4 phages | Non-fastidious aerobic and facultatively anaerobic bacteria | Enterobactermassiliensis strain JC163T |
| Bailey and Coe [122] Rhesus Monkeys | BHI | NA | Non-fastidious aerobic and facultatively anaerobic bacteria | NA |
| EMB | NA | Gram-negative aerobic and facultatively anaerobic bacteria | NA | |
| LBS | NA | Aerobic members of lactobacilli | Lactobacillusspp. | |
| Lei et al. [123] Female mice | GMM | NA | Gut aero-intolerant bacteria | |
| López-Moreno [117] | BHI | Supplemented with Obesogens: BPA, BPS | Anaerobic facultative Firmicutes | Staphylococcus, Bacillusamyloliquefaciens group, Streptococcussalivarius |
| López-Moreno [117] | MRS | Supplemented with Obesogens: BPA, BPS | Lactobacillus, Enterobacteria | Latilactobacillus sakei, Enterococcus faecium |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
López-Moreno, A.; Acuña, I.; Torres-Sánchez, A.; Ruiz-Moreno, Á.; Cerk, K.; Rivas, A.; Suárez, A.; Monteoliva-Sánchez, M.; Aguilera, M. Next Generation Probiotics for Neutralizing Obesogenic Effects: Taxa Culturing Searching Strategies. Nutrients 2021, 13, 1617. https://doi.org/10.3390/nu13051617
López-Moreno A, Acuña I, Torres-Sánchez A, Ruiz-Moreno Á, Cerk K, Rivas A, Suárez A, Monteoliva-Sánchez M, Aguilera M. Next Generation Probiotics for Neutralizing Obesogenic Effects: Taxa Culturing Searching Strategies. Nutrients. 2021; 13(5):1617. https://doi.org/10.3390/nu13051617
Chicago/Turabian StyleLópez-Moreno, Ana, Inmaculada Acuña, Alfonso Torres-Sánchez, Ángel Ruiz-Moreno, Klara Cerk, Ana Rivas, Antonio Suárez, Mercedes Monteoliva-Sánchez, and Margarita Aguilera. 2021. "Next Generation Probiotics for Neutralizing Obesogenic Effects: Taxa Culturing Searching Strategies" Nutrients 13, no. 5: 1617. https://doi.org/10.3390/nu13051617