Janus Dendrimers to Assess the Anti-HCV Activity of Molecules in Cell-Assays
Abstract
:1. Introduction
2. Materials and Methods
2.1. Synthesis and Characterization of the Janus Dendrimers
2.1.1. Synthesis of the Dendrons
2.1.2. Synthesis of the Novel Janus Dendrimers by Cu(I)-Catalyzed Alkyne-Azide Cycloaddition (CuAAC)
2.1.3. Deprotection of Amine Groups
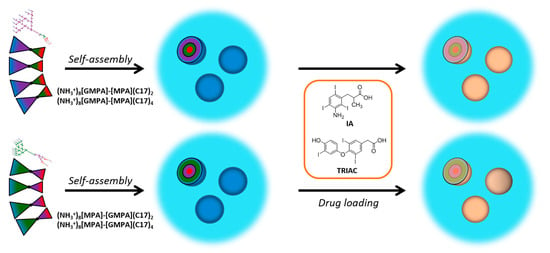
2.2. Formation and Characterization of the Dendrimer Aggregates
2.2.1. Critical Aggregation Concentration (CAC) Determination
2.2.2. Formation and Morphology of the Dendrimer Aggregates
2.3. Formation and Characterization of Drug-Loaded Dendrimer Aggregates
2.3.1. Isothermal Titration Calorimetry (ITC)
2.3.2. Drug Loading
2.4. Antiviral Studies
2.4.1. Cells and Replicon System
2.4.2. Antiviral Assay with Huh 5-2 Cells
2.5. Cell Viability Assay
2.6. Cytometry Assay
3. Results and Discussion
3.1. Synthesis and Chemical Characterization of the Janus Dendrimers
3.2. Preparation and Characterization of the Dendrimer Aggregates
3.2.1. Formation of the Dendrimer Aggregates and Critical Aggregation Concentration (CAC) Determination
3.2.2. Morphological Studies of the Dendrimer Aggregates
3.2.3. Cell Viability Studies in Huh 5-2 Cell Line
3.3. Drug-Loaded Dendrimer Aggregates
3.3.1. Isothermal Titration Calorimetry (ITC) Studies
3.3.2. Drug Loading
3.4. In Vitro Antiviral Activity and Cell Viability of the Dendrimer/Drug Aggregates
3.5. Cellular Internalization Studies by Flow Cytometry Imaging
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Inglese, J.; Johnson, R.L.; Simeonov, A.; Xia, M.; Zheng, W.; Austin, C.P.; Auld, D.S. High-throughput screening assays for the identification of chemical probes. Nat. Chem. Biol. 2007, 3, 466–479. [Google Scholar] [CrossRef] [PubMed]
- Bleicher, K.H.; Böhm, H.J.; Müller, K.; Alanine, A.I. Hit and lead generation: Beyond hi gh-throughput screening. Nat. Rev. Drug Discov. 2003, 2, 369–378. [Google Scholar] [CrossRef] [PubMed]
- Noah, J. New developments and emerging trends in high-throughput screening methods for lead compound identification. Int. J. High Throughput Screen. 2010, 1, 141–149. [Google Scholar] [CrossRef] [Green Version]
- García-Serradilla, M.; Risco, C.; Pacheco, B. Drug repurposing for new, efficient, broad spectrum antivirals. Virus Res. 2020, 264, 22–31. [Google Scholar] [CrossRef] [PubMed]
- Koizumi, Y.; Ohashi, H.; Nakajima, S.; Tanaka, Y.; Wakita, T.; Perelson, A.S.; Iwami, S.; Watashi, K. Quantifying antiviral activity optimizes drug combinations against hepatitis C virus infection. Proc. Natl. Acad. Sci. USA 2017, 114, 1922–1927. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abian, O.; Vega, S.; Sancho, J.; Velazquez-Campoy, A. Allosteric Inhibitors of the NS3 Protease from the Hepatitis C Virus. PLoS ONE 2013, 8, e69773. [Google Scholar] [CrossRef] [PubMed]
- Claveria-Gimeno, R.; Vega, S.; Grazu, V.; de la Fuente, J.M.; Lanas, A.; Velazquez-Campoy, A.; Abian, O. Rescuing compound bioactivity in a secondary cell-based screening by using γ-cyclodextrin as a molecular carrier. Int. J. Nanomed. 2015, 10, 2249–2259. [Google Scholar] [CrossRef] [Green Version]
- Torchilin, V.P. Multifunctional nanocarriers. Adv. Drug Deliv. Rev. 2006, 58, 1532–1555. [Google Scholar] [CrossRef] [PubMed]
- Patil, A.; Mishra, V.; Thakur, S.; Riyaz, B.; Kaur, A.; Khursheed, R.; Patil, K.; Sathe, B. Nanotechnology Derived Nanotools in Biomedical Perspectives: An Update. Curr. Nanosci. 2019, 15, 137–146. [Google Scholar] [CrossRef]
- Su, C.; Liu, Y.; Li, R.; Wu, W.; Fawcett, J.P.; Gu, J. Absorption, distribution, metabolism and excretion of the biomaterials used in Nanocarrier drug delivery systems. Adv. Drug Deliv. Rev. 2019, 143, 97–114. [Google Scholar] [CrossRef]
- Sherje, A.P.; Jadhav, M.; Dravyakar, B.R.; Kadam, D. Dendrimers: A versatile nanocarrier for drug delivery and targeting. Int. J. Pharm. 2018, 548, 707–720. [Google Scholar] [CrossRef]
- Sandoval-Yañez, C.; Rodriguez, C.C. Dendrimers: Amazing platforms for bioactive molecule delivery systems. Materials 2020, 13, 570. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mendes, L.P.; Pan, J.; Torchilin, V.P. Dendrimers as nanocarriers for nucleic acid and drug delivery in cancer therapy. Molecules 2017, 22, 1401. [Google Scholar] [CrossRef] [Green Version]
- Chahal, J.S.; Khan, O.F.; Cooper, C.L.; Mcpartlan, J.S.; Tsosie, J.K.; Tilley, L.D.; Sidik, S.M.; Lourido, S.; Langer, R.; Bavari, S.; et al. Dendrimer-RNA nanoparticles generate protective immunity against lethal Ebola, H1N1 influenza, and Toxoplasma gondii challenges with a single dose. Proc. Natl. Acad. Sci. USA 2016, 113, E4133–E4142. [Google Scholar] [CrossRef] [Green Version]
- Bahadoran, A.; Moeini, H.; Bejo, M.H.; Hussein, M.Z.; Omar, A.R. Development of tat-conjugated dendrimer for transdermal DNA vaccine delivery. J. Pharm. Pharm. Sci. 2016, 19, 325–338. [Google Scholar] [CrossRef] [Green Version]
- Mhlwatika, Z.; Aderibigbe, B.A. Application of dendrimers for the treatment of infectious diseases. Molecules 2018, 23, 2205. [Google Scholar] [CrossRef] [Green Version]
- Mintzer, M.A.; Dane, E.L.; O’Toole, G.A.; Grinstaff, M.W. Exploiting dendrimer multivalency to combat emerging and re-emerging infectious diseases. Mol. Pharm. 2012, 9, 342–354. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Gitsov, I. Thermosensitive Amphiphilic Janus Dendrimers with Embedded Metal Binding Sites. Synthesis and Self-Assembly. Macromolecules 2018, 51, 5085–5100. [Google Scholar] [CrossRef]
- Buzzacchera, I.; Xiao, Q.; Han, H.; Rahimi, K.; Li, S.; Kostina, N.Y.; Toebes, B.J.; Wilner, S.E.; Möller, M.; Rodriguez-Emmenegger, C.; et al. Screening Libraries of Amphiphilic Janus Dendrimers Based on Natural Phenolic Acids to Discover Monodisperse Unilamellar Dendrimersomes. Biomacromolecules 2019, 20, 712–727. [Google Scholar] [CrossRef]
- Sherman, S.E.; Xiao, Q.; Percec, V. Mimicking complex biological membranes and their programmable glycan ligands with dendrimersomes and glycodendrimersomes. Chem. Rev. 2017, 117, 6538–6631. [Google Scholar] [CrossRef]
- Caminade, A.M.; Laurent, R.; Delavaux-Nicot, B.; Majoral, J.P. “Janus” dendrimers: Syntheses and properties. New J. Chem. 2012, 36, 217–226. [Google Scholar] [CrossRef]
- Sikwal, D.R.; Kalhapure, R.S.; Govender, T. An emerging class of amphiphilic dendrimers for pharmaceutical and biomedical applications: Janus amphiphilic dendrimers. Eur. J. Pharm. Sci. 2017, 97, 113–134. [Google Scholar] [CrossRef] [PubMed]
- Taabache, S.; Bertin, A. Vesicles from amphiphilic dumbbells and janus dendrimers: Bioinspired self-assembled structures for biomedical applications. Polymers 2017, 9, 280. [Google Scholar] [CrossRef] [Green Version]
- Najafi, F.; Salami-Kalajahi, M.; Roghani-Mamaqani, H. Synthesis of amphiphilic Janus dendrimer and its application in improvement of hydrophobic drugs solubility in aqueous media. Eur. Polym. J. 2020, 134, 109804. [Google Scholar] [CrossRef]
- Lancelot, A.; Clavería-Gimeno, R.; Velázquez-Campoy, A.; Abian, O.; Serrano, J.L.; Sierra, T. Nanostructures based on ammonium-terminated amphiphilic Janus dendrimers as camptothecin carriers with antiviral activity. Eur. Polym. J. 2017, 90, 136–149. [Google Scholar] [CrossRef]
- Lancelot, A.; González-Pastor, R.; Clavería-Gimeno, R.; Romero, P.; Abian, O.; Martín-Duque, P.; Serrano, J.L.; Sierra, T. Cationic poly(ester amide) dendrimers: Alluring materials for biomedical applications. J. Mater. Chem. B 2018, 6, 3956–3968. [Google Scholar] [CrossRef] [PubMed]
- Fedeli, E.; Lancelot, A.; Serrano, J.L.; Calvo, P.; Sierra, T. Self-assembling amphiphilic Janus dendrimers: Mesomorphic properties and aggregation in water. New J. Chem. 2015, 39, 1960–1967. [Google Scholar] [CrossRef] [Green Version]
- Movellan, J.; Urbán, P.; Moles, E.; de la Fuente, J.M.; Sierra, T.; Serrano, J.L.; Fernàndez-Busquets, X. Amphiphilic dendritic derivatives as nanocarriers for the targeted delivery of antimalarial drugs. Biomaterials 2014, 35, 7940–7950. [Google Scholar] [CrossRef]
- Stenström, P.; Hjorth, E.; Zhang, Y.; Andrén, O.C.J.; Guette-Marquet, S.; Schultzberg, M.; Malkoch, M. Synthesis and in Vitro Evaluation of Monodisperse Amino-Functional Polyester Dendrimers with Rapid Degradability and Antibacterial Properties. Biomacromolecules 2017, 18, 4323–4330. [Google Scholar] [CrossRef]
- Stuart, M.C.A.; Van De Pas, J.C.; Engberts, J.B.F.N. The use of Nile Red to monitor the aggregation behavior in ternary surfactant-water-organic solvent systems. J. Phys. Org. Chem. 2005, 18, 929–934. [Google Scholar] [CrossRef] [Green Version]
- Vega, S.; Abian, O.; Velazquez-Campoy, A. A unified framework based on the binding polynomial for characterizing biological systems by isothermal titration calorimetry. Methods 2015, 76, 99–115. [Google Scholar] [CrossRef] [Green Version]
- Courcambeck, J.; Bouzidi, M.; Perbost, R.; Jouirou, B.; Amrani, N.; Cacoub, P.; Pèpe, G.; Sabatier, J.M.; Halfon, P. Resistance of hepatitis C virus to NS3-4A protease inhibitors: Mechanisms of drug resistance induced by R155Q, A156T, D168A and D168V mutations. Antivir. Ther. 2006, 11, 847–855. [Google Scholar] [PubMed]
- Susser, S.; Vermehren, J.; Forestier, N.; Welker, M.W.; Grigorian, N.; Füller, C.; Perner, D.; Zeuzem, S.; Sarrazin, C. Analysis of long-term persistence of resistance mutations within the hepatitis C virus NS3 protease after treatment with telaprevir or boceprevir. J. Clin. Virol. 2011, 52, 321–327. [Google Scholar] [CrossRef]
- Blight, K.J.; McKeating, J.A.; Rice, C.M. Highly Permissive Cell Lines for Subgenomic and Genomic Hepatitis C Virus RNA Replication. J. Virol. 2002, 76, 13001–13014. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lohmann, V.; Körner, F.; Koch, J.O.; Herian, U.; Theilmann, L.; Bartenschlager, R. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 1999, 285, 110–113. [Google Scholar] [CrossRef] [Green Version]
- Urbani, A.; Bazzo, R.; Nardi, M.C.; Cicero, D.O.; De Francesco, R.; Steinkühler, C.; Barbato, G. The metal binding site of the hepatitis C virus NS3 protease: A spectroscopic investigation. J. Biol. Chem. 1998, 273, 18760–18769. [Google Scholar] [CrossRef] [Green Version]
- Franc, G.; Kakkar, A.K. “Click” methodologies: Efficient, simple and greener routes to design dendrimers. Chem. Soc. Rev. 2010, 39, 1536–1544. [Google Scholar] [CrossRef]
- Fedeli, E.; Lancelot, A.; Dominguez, J.M.; Serrano, J.L.; Calvo, P.; Sierra, T. Self-assembling hybrid linear-dendritic block copolymers: The design of nano-carriers for lipophilic antitumoral drugs. Nanomaterials 2019, 9, 161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Del Barrio, J.; Oriol, L.; Sánchez, C.; Serrano, J.L.; Di Cicco, A.; Keller, P.; Li, M.H. Self-assembly of linear-dendritic diblock copolymers: From nanofibers to polymersomes. J. Am. Chem. Soc. 2010, 132, 3762–3769. [Google Scholar] [CrossRef]
- Liu, X.; Gitsov, I. Nonionic amphiphilic linear dendritic block copolymers. solvent-induced self-Assembly and morphology tuning. Macromolecules 2019, 52, 5563–5573. [Google Scholar] [CrossRef]
- Jiménez-Pardo, I.; González-Pastor, R.; Lancelot, A.; Clavería-Gimeno, R.; Velázquez-Campoy, A.; Abian, O.; Ros, M.B.; Sierra, T. Shell Cross-Linked Polymeric Micelles as Camptothecin Nanocarriers for Anti-HCV Therapy. Macromol. Biosci. 2015, 15, 1381–1391. [Google Scholar] [CrossRef] [Green Version]










| Dendrimer | IA | TRIAC | |||||
|---|---|---|---|---|---|---|---|
| Lc a (%) | CAC (M) | DH (nm) | [IA] (μM) | DLCc (mol/mol) | [TRIAC] (μM) | DLCc (mol/mol) | |
| (NH3+)8[GMPA]-[MPA](C17)2 | 9.7 | 1.1·10−5 | 9 ± 1 | 680 ± 172 | 1.86 | 563 ± 257 | 1.65 |
| (NH3+)8[GMPA]-[MPA](C17)4 | 14.8 | 6.9·10−6 | 24 ± 1 | 321 ± 119 | 1.44 | 505 ± 238 | 1.59 |
| (NH3+)8[MPA]-[GMPA](C17)2 | 10.7 | 8.1·10−6 | 17 ± 3 | 641 ± 207 | 1.78 | 583 ± 167 | 1.81 |
| (NH3+)8[MPA]-[GMPA](C17)4 | 14.8 | 1.2·10−5 | 34 ± 5 b | 35 ± 16 | 0.19 | 70 ± 49 | 0.29 |
| Ka | ΔG | ΔH | −T·ΔS | ||
|---|---|---|---|---|---|
| (NH3+)8[GMPA]-[MPA](C17)2 | IA | 6.3 × 103 | −5.2 | −1.1 | −4.1 |
| TRIAC | 1.0 × 105 | −6.8 | −6.0 | −0.8 | |
| (NH3+)8[GMPA]-[MPA](C17)4 | IA | 9.2 × 104 | −6.8 | −0.4 | −6.4 |
| TRIAC | 6.1 × 105 | −7.9 | −0.4 | −7.5 | |
| (NH3+)8[MPA]-[GMPA](C17)2 | IA | 4.8 × 104 | −6.4 | −0.1 | −6.3 |
| TRIAC | 1.5 × 105 | −7.1 | −1.3 | −5.8 | |
| (NH3+)8[MPA]-[GMPA](C17)4 | IA | 1.1 × 106 | −8.2 | 0.7 | −8.9 |
| TRIAC | 8.8 × 104 | −6.7 | −15.8 | 9.1 |
| [IA] (μM) | [TRIAC] (μM) | |||
|---|---|---|---|---|
| CC50 | EC40 | CC50 | EC40 | |
| (NH3+)8[GMPA]-[MPA](C17)2 | >160 | 2.5–5 | >63 | 1 |
| (NH3+)8[GMPA]-[MPA](C17)4 | >80 | 2.5 | >67 | 1 |
| (NH3+)8[MPA]-[GMPA](C17)2 | >99 | >99 | >61 | 30–60 |
| free drug | 300 | 175 | 188 | 20–40 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
San Anselmo, M.; Lancelot, A.; Egido, J.E.; Clavería-Gimeno, R.; Casanova, Á.; Serrano, J.L.; Hernández-Ainsa, S.; Abian, O.; Sierra, T. Janus Dendrimers to Assess the Anti-HCV Activity of Molecules in Cell-Assays. Pharmaceutics 2020, 12, 1062. https://doi.org/10.3390/pharmaceutics12111062
San Anselmo M, Lancelot A, Egido JE, Clavería-Gimeno R, Casanova Á, Serrano JL, Hernández-Ainsa S, Abian O, Sierra T. Janus Dendrimers to Assess the Anti-HCV Activity of Molecules in Cell-Assays. Pharmaceutics. 2020; 12(11):1062. https://doi.org/10.3390/pharmaceutics12111062
Chicago/Turabian StyleSan Anselmo, María, Alexandre Lancelot, Julia E. Egido, Rafael Clavería-Gimeno, Álvaro Casanova, José Luis Serrano, Silvia Hernández-Ainsa, Olga Abian, and Teresa Sierra. 2020. "Janus Dendrimers to Assess the Anti-HCV Activity of Molecules in Cell-Assays" Pharmaceutics 12, no. 11: 1062. https://doi.org/10.3390/pharmaceutics12111062