Characterization and Therapeutic Effect of a pH Stimuli Responsive Polymeric Nanoformulation for Controlled Drug Release
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Synthesis of Aminomethyl Polystyrene Nanoparticles (NPs)
2.3. PEGylation of NPs
2.4. Preparation of pH Responsive Therapeutic Polymeric Nanodevice: DOX-NPs (7)
2.5. Characterization of DOX-NPs (7)
2.5.1. Nanoparticle Size Distribution, Zeta Potential, and Morphology
2.5.2. Determination of Nanoparticle Concentration by Spectrophotometry
2.5.3. Amino Quantification of Nanoparticles
2.5.4. Evaluation of Drug-Loading Efficiency
2.5.5. Drug Release Profile
2.6. Cell Cultures
2.7. Nanofection of Cancer Cell Lines
2.8. Cell Viability
2.9. Determination of DNA Damage in Cancer Cells by Immunostaining of Phospho-H2A.X Foci
2.10. Statistical Analysis
3. Results and Discussion
3.1. Preparation of DOX-NPs (7)
Characterization of Drug-Loaded Nanoparticles (DOX-NPs) (7)
3.2. Evaluation of the Efficiency of Cellular Uptake of DOX-NP (7)
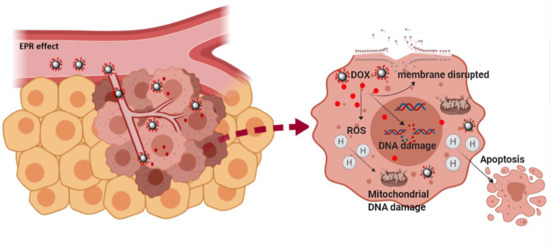
3.3. Evaluation of the Therapeutic Capacity of the DOX-NP (7)
3.4. Analysis of DOX-NPs (7)-Induced Genotoxic Effect in Cancer Cells
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Blau, R.; Krivitsky, A.; Epshtein, Y.; Satchi-Fainaro, R. Are nanotheranostics and nanodiagnostics-guided drug delivery stepping stones towards precision medicine? Drug Resist. Updat. 2016, 27, 39–58. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Roy, I.; Yang, C.; Prasad, P.N. Nanochemistry and Nanomedicine for Nanoparticle-based Diagnostics and Therapy. Chem. Rev. 2016, 116, 2826–2885. [Google Scholar] [CrossRef] [PubMed]
- Cagel, M.; Grotz, E.; Bernabeu, E.; Moretton, M.A.; Chiappetta, D.A. Doxorubicin: Nanotechnological overviews from bench to bedside. Drug Discov. Today 2017, 22, 270–281. [Google Scholar] [CrossRef] [PubMed]
- El-Say, K.M.; El-Sawy, H.S. Polymeric nanoparticles: Promising platform for drug delivery. Int. J. Pharm. 2017, 528, 675–691. [Google Scholar] [CrossRef]
- Ekladious, I.; Colson, Y.L.; Grinstaff, M.W. Polymer–drug conjugate therapeutics: advances, insights and prospects. Nat. Rev. Drug Discov. 2019, 18, 273–294. [Google Scholar] [CrossRef]
- Chenthamara, D.; Subramaniam, S.; Ramakrishnan, S.G.; Krishnaswamy, S.; Essa, M.M.; Lin, F.H.; Qoronfleh, M.W. Therapeutic efficacy of nanoparticles and routes of administration. Biomater. Res. 2019, 23, 1–29. [Google Scholar] [CrossRef]
- Park, J.; Choi, Y.; Chang, H.; Um, W.; Ryu, J.H.; Kwon, I.C. Alliance with EPR effect: Combined strategies to improve the EPR effect in the tumor microenvironment. Theranostics 2019, 9, 8073–8090. [Google Scholar] [CrossRef]
- Katz, J.S.; Burdick, J.A. Light-Responsive Biomaterials: Development and Applications. Macromol. Biosci. 2010, 10, 339–348. [Google Scholar] [CrossRef]
- Karimi, M.; Sahandi, Z.P.; Ghasemi, A.; Amiri, M.; Bahrami, M.; Malekzad, H.; Ghahramanzadeh Asl, H.; Mahdieh, Z.; Bozorgomid, M.; Ghasemi, A.; et al. Temperature-Responsive Smart Nanocarriers for Delivery of Therapeutic Agents: Applications and Recent Advances. ACS Appl. Mater. Interfaces 2016, 8, 21107–21133. [Google Scholar] [CrossRef]
- Zhao, Y.; Luo, Z.; Li, M.; Qu, Q.; Ma, X.; Yu, S.-H.; Zhao, Y. A Preloaded Amorphous Calcium Carbonate/Doxorubicin@Silica Nanoreactor for pH-Responsive Delivery of an Anticancer Drug. Angew. Chem. Int. Ed. 2015, 54, 919–922. [Google Scholar] [CrossRef]
- Corbet, C.; Feron, O. Tumour acidosis: From the passenger to the driver’s seat. Nat. Rev. Cancer 2017, 17, 577–593. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Zhang, Y.S.; Pang, B.; Hyun, D.C.; Yang, M.; Xia, Y. Engineered Nanoparticles for Drug Delivery in Cancer Therapy. Angew. Chem. Int. Ed. 2014, 12320–12364. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yang, C.; Wang, W.; Liu, J.; Liu, Q.; Huang, F.; Chu, L.; Gao, H.; Li, C.; Kong, D.; et al. Co-delivery of doxorubicin and curcumin by pH-sensitive prodrug nanoparticle for combination therapy of cancer. Sci. Rep. 2016, 6, 21225. [Google Scholar] [CrossRef] [PubMed]
- Cui, T.; Liang, J.-J.; Chen, H.; Geng, D.-D.; Jiao, L.; Yang, J.-Y.; Qian, H.; Zhang, C.; Ding, Y. Performance of Doxorubicin-Conjugated Gold Nanoparticles: Regulation of Drug Location. ACS Appl. Mater. Interfaces 2017, 9, 8569–8580. [Google Scholar] [CrossRef]
- Thorn, C.; Oshiro, C.; Marsh, S.; Hernandez-Boussard, T.; McLeod, H.; Klein, T.; Altman, R. Doxorubicin pathways:pharmacodynamics and adverse effects. Pharmacogn. Genomics 2012, 21, 440–446. [Google Scholar] [CrossRef]
- Barenholz, Y.C. Doxil®—The first FDA-approved nano-drug: Lessons learned. J. Control. Release 2012, 160, 117–134. [Google Scholar] [CrossRef]
- Huang, Y.; Yan, J.; Peng, S.; Tang, Z.; Tan, C.; Ling, J.; Lin, W.; Lin, X.; Zu, X.; Yi, G. pH/reduction dual-stimuli-responsive cross-linked micelles based on multi-functional amphiphilic star copolymer: Synthesis and controlled anti-cancer drug release. Polymers 2020, 12, 82. [Google Scholar] [CrossRef]
- Raposo, C.D.; Costa, R.; Petrova, K.T.; Brito, C.; Scotti, M.T.; Cardoso, M.M. Development of novel galactosylated PLGA nanoparticles for hepatocyte targeting using molecular modelling. Polymers 2020, 12, 94. [Google Scholar] [CrossRef]
- Gibbens-Bandala, B.; Morales-Avila, E.; Ferro-Flores, G.; Santos-Cuevas, C.; Luna-Gutiérrez, M.; Ramírez-Nava, G.; Ocampo-García, B. Synthesis and evaluation of 177Lu-DOTA-DN(PTX)-BN for selective and concomitant radio and drug-therapeutic effect on breast cancer cells. Polymers 2019, 11, 1572. [Google Scholar] [CrossRef]
- Altea-Manzano, P.; Unciti-Broceta, J.D.; Cano-Cortes, V.; Ruiz-Blas, M.P.; Valero-Griñan, T.; Diaz-Mochon, J.J.; Sanchez-Martin, R. Tracking cell proliferation using a nanotechnology-based approach. Nanomedicine 2017, 12, 1591–1605. [Google Scholar] [CrossRef]
- Valero, T.; Delgado-González, A.; Unciti-Broceta, J.D.; Cano-Cortés, V.; Pérez-López, A.M.; Unciti-Broceta, A.; Sánchez Martín, R.M. Drug “Clicking” on Cell-Penetrating Fluorescent Nanoparticles for In Cellulo Chemical Proteomics. Bioconjug. Chem. 2018, 29, 3154–3160. [Google Scholar] [CrossRef] [PubMed]
- Delgado-Gonzalez, A.; Garcia-Fernandez, E.; Valero, T.; Cano-Cortes, M.V.; Ruedas-Rama, M.J.; Unciti-Broceta, A.; Sanchez-Martin, R.M.; Diaz-Mochon, J.J.; Orte, A. Metallofluorescent Nanoparticles for Multimodal Applications. ACS Omega 2018, 3, 144–153. [Google Scholar] [CrossRef] [PubMed]
- Alexander, L.M.; Pernagallo, S.; Livigni, A.; Sánchez-Martín, R.M.; Brickman, J.M.; Bradley, M. Investigation of microsphere-mediated cellular delivery by chemical, microscopic and gene expression analysis. Mol. Biosyst. 2010, 6, 399–409. [Google Scholar] [CrossRef]
- Pietrovito, L.; Cano-Cortés, V.; Gamberi, T.; Magherini, F.; Bianchi, L.; Bini, L.; Sánchez-Martín, R.M.; Fasano, M.; Modesti, A. Cellular response to empty and palladium-conjugated amino-polystyrene nanospheres uptake: A proteomic study. Proteomics 2015, 15, 34–43. [Google Scholar] [CrossRef]
- Unciti-Broceta, A.; Johansson, E.M.V.; Yusop, M.R.; Sánchez-Martín, R.M.; Bradley, M. Synthesis of polystyrene microspheres and functionalization with Pd0 nanoparticles to perform bioorthogonal organometallic chemistry in living cells. Nat. Protoc. 2012, 7, 1207–1218. [Google Scholar] [CrossRef] [PubMed]
- Unciti-Broceta, J.D.; Cano-Cortés, V.; Altea-Manzano, P.; Pernagallo, S.; Díaz-Mochón, J.J.; Sánchez-Martín, R.M. Number of nanoparticles per cell through a spectrophotometric method-A key parameter to assess nanoparticle-based cellular assays. Sci. Rep. 2015, 5, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Fields, G.B.; Noble, R.L. Solid phase peptide synthesis utilizing 9-fluorenylmethoxycarbonyl amino acids. Int. J. Pept. Protein Res. 1990, 35, 161–214. [Google Scholar] [CrossRef]
- Francesco, C.; Larissa, L.; Antonio, G.; Joanna, C.; Alexey, P.; Alexandra, D.; Katarzyna, S.-K.; Alessandro, P.; Sergey, P.; Barlev, N.A. Specific Drug Delivery to Cancer Cells with Double-Imprinted Nanoparticles against Epidermal Growth Factor Receptor. Nano Lett. 2018, 18, 4641–4646. [Google Scholar]
- Arcamone, F.; Cassinelli, G.; Fantini, G.; Grein, A.; Orezzi, P.; Pol, C.; Spalla, C. Adriamycin, 14-hydroxydaimomycin, a new antitumor antibiotic fromS. Peucetius var.caesius. Biotechnol. Bioeng. 1969, 11, 1101–1110. [Google Scholar] [CrossRef]
- Cortés-Funes, H.; Coronado, C. Role of anthracyclines in the era of targeted therapy. Cardiovasc. Toxicol. 2007, 7, 56–60. [Google Scholar] [CrossRef]
- Weiss, R.B. The anthracyclines: Will we ever find a better doxorubicin? Semin. Oncol. 1992, 19, 670–686. [Google Scholar] [PubMed]
- No Manchun, S.; Dass, C.R.; Sriamornsak, P. Targeted Therapy for Cancer Using Ph-Responsive Nanocarrier Systems. Life Sci. 2012, 90, 381–387. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.; Wilson, I.; Orton, T.; Pognan, F. Investigation of the Alamar Blue (resazurin) fluorescent dye for the assessment of mammalian cell cytotoxicity. Eur. J. Biochem. 2000, 267, 5421–5426. [Google Scholar]
- Gewirtz, D.A. A critical evaluation of the mechanisms of action proposed for the antitumor effects of the anthracycline antibiotics adriamycin and daunorubicin. Biochem. Pharmacol. 1999, 57, 727–741. [Google Scholar] [CrossRef]





© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cano-Cortes, M.V.; Laz-Ruiz, J.A.; Diaz-Mochon, J.J.; Sanchez-Martin, R.M. Characterization and Therapeutic Effect of a pH Stimuli Responsive Polymeric Nanoformulation for Controlled Drug Release. Polymers 2020, 12, 1265. https://doi.org/10.3390/polym12061265
Cano-Cortes MV, Laz-Ruiz JA, Diaz-Mochon JJ, Sanchez-Martin RM. Characterization and Therapeutic Effect of a pH Stimuli Responsive Polymeric Nanoformulation for Controlled Drug Release. Polymers. 2020; 12(6):1265. https://doi.org/10.3390/polym12061265
Chicago/Turabian StyleCano-Cortes, Maria Victoria, Jose Antonio Laz-Ruiz, Juan Jose Diaz-Mochon, and Rosario Maria Sanchez-Martin. 2020. "Characterization and Therapeutic Effect of a pH Stimuli Responsive Polymeric Nanoformulation for Controlled Drug Release" Polymers 12, no. 6: 1265. https://doi.org/10.3390/polym12061265