Published online Oct 15, 2021. doi: 10.4251/wjgo.v13.i10.1263
Peer-review started: February 21, 2021
First decision: May 8, 2021
Revised: May 18, 2021
Accepted: August 27, 2021
Article in press: August 27, 2021
Published online: October 15, 2021
Pancreatic cancer (PC) continues to pose a major clinical challenge. There has been little improvement in patient survival over the past few decades, and it is projected to become the second leading cause of cancer mortality by 2030. The dismal 5-year survival rate of less than 10% after the diagnosis is attributable to the lack of early symptoms, the absence of specific biomarkers for an early diagnosis, and the inadequacy of available chemotherapies. Most patients are diagnosed when the disease has already metastasized and cannot be treated. Cancer interception is vital, actively intervening in the malignization process before the development of a full-blown advanced tumor. An early diagnosis of PC has a dramatic impact on the survival of patients, and improved techniques are urgently needed to detect and evaluate this disease at an early stage. It is difficult to obtain tissue biopsies from the pancreas due to its anatomical position; however, liquid biopsies are readily available and can provide useful information for the diagnosis, prognosis, stratification, and follow-up of patients with PC and for the design of individually tailored treatments. The aim of this review was to provide an update of the latest advances in knowledge on the application of carbohydrates, proteins, cell-free nucleic acids, circulating tumor cells, metabo
Core Tip: Pancreatic cancer (PC) is still an especially lethal malignancy, with a 5-year survival rate below 10%. Liquid biopsies are a readily-available source of biomarkers to determine the situation of patients. This review summarizes the most recent published findings on the diagnostic, prognostic, and predictive potential for PC of biomarkers identified in liquid biopsies, discussing the strengths, limitations, and drawbacks of their clinical application. There remains a need to validate and verify the clinical value of liquid biopsies for PC in large-scale human trials using appropriate controls.
- Citation: Perales S, Torres C, Jimenez-Luna C, Prados J, Martinez-Galan J, Sanchez-Manas JM, Caba O. Liquid biopsy approach to pancreatic cancer. World J Gastrointest Oncol 2021; 13(10): 1263-1287
- URL: https://www.wjgnet.com/1948-5204/full/v13/i10/1263.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v13.i10.1263
Despite recent advances in understanding the molecular events underlying pancreatic cancer (PC), it remains one of the deadliest malignancies[1]. In the absence of novel diagnostic methods and/or treatments, PC is expected to become the second leading cause of cancer-related deaths by 2030[2]. Complete surgical resection is a potentially curative treatment, but 80%-85% of patients present with non-resectable disease at their diagnosis due to local invasion or distant metastasis[3]. An early diagnosis is crucial but is highly challenging due to the lack of specific symptoms during initial stages of the disease. The etiology of PC is not well understood, and new prospective studies are needed to fully elucidate related factors[4]. The development of diagnostic, prognostic, and predictive biomarkers is urgently needed to improve the clinical management of PC[5].
An ideal tumor biomarker is both specific, i.e., exclusive to the disease in question, and sensitive, detecting early cancer development and identifying all cases. It should have positive predictive value, enabling detection in the general population during very early stages, and a long lead time, sufficient to allow modification of the natural course of the disease. In addition, it should correlate with tumor stage and allow the monitoring of biomarker levels to detect early relapse. Finally, the biomarker should be easy to collect and involve a simple and economically feasible test[6]. Ideally, a diagnostic biomarker allows PC to be detected at an early stage, a prognostic biomarker provides information on the disease in untreated individuals, and a predictive biomarker yields information for personalizing treatment to obtain the best therapeutic response and deliver optimal patient care[7].
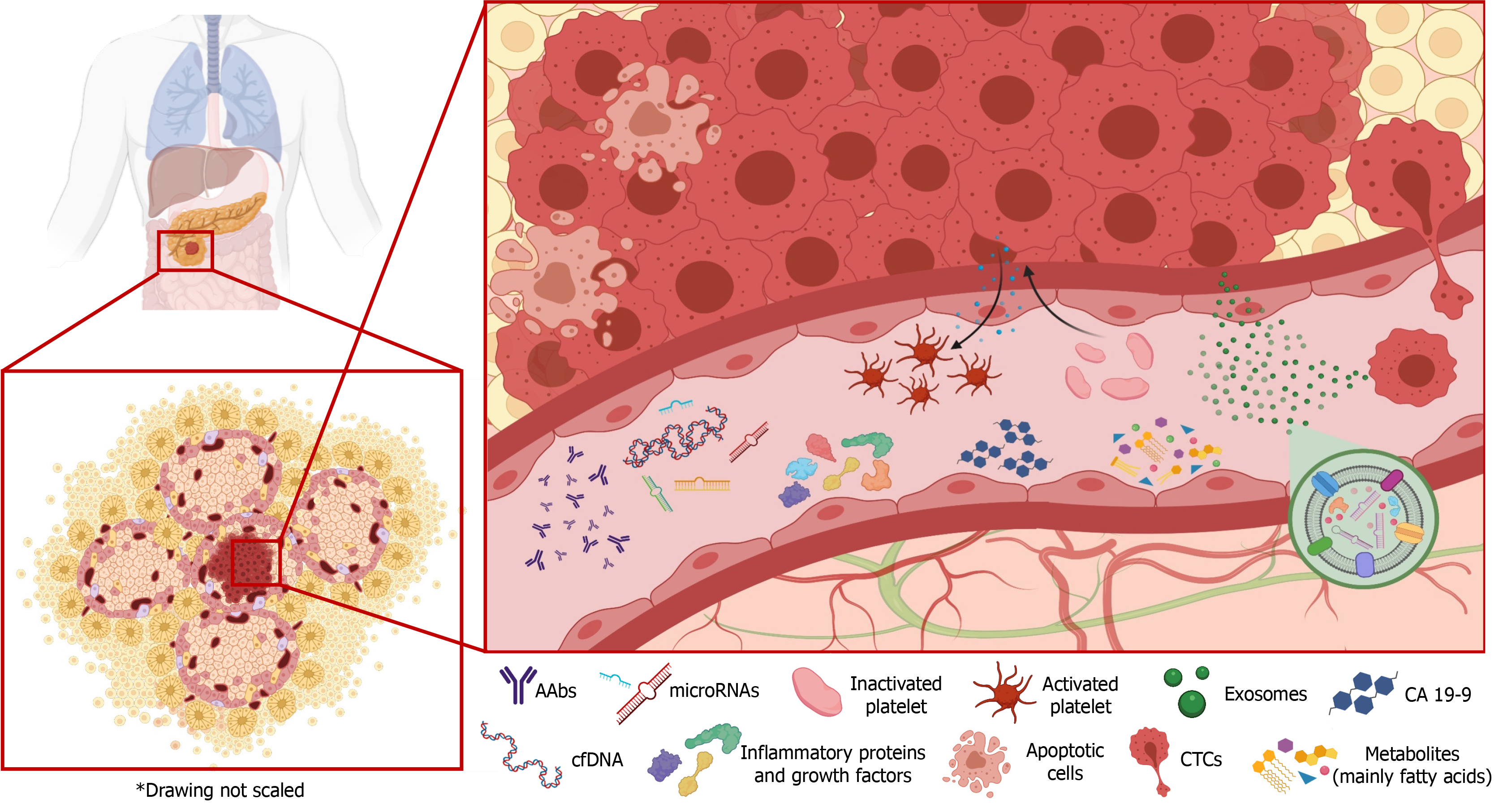
Liquid biopsy is a promising non-invasive matrix that has proven useful for the early diagnosis of various types of cancer. It offers a novel tool for monitoring tumor stage and the response to treatment throughout disease progression[8]. However, unlike in some other cancers, the clinical applicability and reliability of liquid biopsy results have yet to be established in PC[9]. The aim of this review was to provide an update on recent advances in the development of liquid biopsy biomarkers for PC and evaluate their potential clinical usefulness (Figure 1).
Carbohydrate antigen (CA) 19-9, an epitope of sialylated Lewis antigen, appears elevated in PC[10], and there have been numerous studies on its usefulness for screening, diagnostic, prognostic, and predictive purposes and for assessing resectability[11]. It has been documented as an early detector of PC[12], and it is the only biomarker approved for monitoring its progression and the therapeutic response[13]. However, some shortcomings have been reported, including poor sensitivity (69%-98%) and specificity (46%-98%) for PC diagnosis[14]. Furthermore, it is not tumor-specific, given that elevated concentrations of CA 19-9 have also been observed in other gastrointestinal malignancies (e.g., colorectal cancer and gastric cancer) and benign diseases (e.g., chronic pancreatitis (CP) and acute cholangitis)[15]. Finally, CA 19-9 is not expressed by around 10% of Caucasian individuals, even those with large PC tumors, because of their possession of the Lewis a-/b- genotype[16].
Other carbohydrate markers that have been related to PC include CA 50, CA 72-4, CA 125, and CA 242. However, these biomarkers have been little used in the clinical management of PC due to their low sensitivity and specificity. CA 242 contains a different sialylated carbohydrate type I chain to that in CA 19-9 and was found to offer lower sensitivity but higher specificity for PC[17]; however, elevated concentrations of CA 242 are found in other types of cancer (e.g., cervical and lymphoma), in type 2 diabetes mellitus, and in coronary heart disease, suggesting an association between this biomarker and the presence of chronic disease in general[18]. CA 125, also known as mucin 16, has the same drawback, being most frequently used in the diagnosis of ovarian cancer[19], and altered CA 125 concentrations have been observed in a wide range of diseases, including heart failure[20], pulmonary disorders[21], deep endometriosis[22], and extrapulmonary tuberculosis[23].
There has been little published research on the clinical relevance of carcinoembryonic antigen (CEA) in PC, which has been widely used to diagnose colon cancer[24] and also appears elevated in around 60% of patients with PC. Its sensitivity to diagnose PC is only around 37%, and elevated concentrations have been observed in patients with nonspecific colitis and in heavy smokers. For this reason, CEA has been less frequently utilized in comparison to CA 19–9. However, it may be useful to evaluate the effectiveness of treatment and to detect recurrence in patients with a diagnosis of PC[25]. It has been described as a better predictive biomarker than CA 19-9 in metastatic and locally advanced PC[26] but a worse predictive biomarker for survival and early recurrence[27]. Finally, alongside other carbohydrate biomarkers, CEA may be of value in the clinical management of Lewis-negative patients, achieving high specificity (98.0%) for the diagnosis of PC and being related to the metastatic status and response to therapy[28].
None of the above markers are sufficiently accurate when applied alone, and recent studies have focused on their combined use in panels. Notably, Zhang et al[29] carried out a meta-analysis on the capacity of CA 19-9, CA 242, and CEA to diagnose PC, selecting 21 studies that included 3497 participants. They obtained higher sensitivity with the combination of CA 19-9 plus CA 242 (89.95%) than with the separate application of CA 19-9 or CA 242.
More recently, Deng et al[30] correlated CA 19–9, CA 125, and CEA with multiple clinical factors in PC, relating their baseline levels to the primary tumor site, number of organ metastases, and liver metastases. They reported that the baseline neutrophil count, lactate dehydrogenase enzyme concentration, and CA 19–9 and CA 125 Levels were associated with overall survival (OS) and proposed them as potential prognostic factors.
This combination of CA 19-9, CA 125, and CEA has also been described as useful in the surgical setting. After a study of more than 1000 patients with PC, Xu et al[31] proposed the preoperative serum signature of CA 19-9 ≥ 1000 U/mL, CA 125+, and CEA+ to rule out pancreatectomy, because resected patients with this signature showed no survival advantage over those with locally advanced disease who did not undergo pancreatectomy.
In summary, despite the modest results obtained with CA 19-9 and its poor specificity, it remains the standard against which any new biomarker is compared for the clinical management of PC. Currently, no single carbohydrate marker or combination of markers appears to be useful for this purpose. The main limitations of research to date are the lack of standardization and the absence of validation studies in larger cohorts[32].
Metabolomics is a powerful technology that analyzes the concentrations of low-molecular-weight molecules, also known as metabolites. An increase in sensitivity, resolution, reproducibility, and coverage has been achieved over the past few years through its combination with improved analytical techniques such as high-resolution mass spectrometry (HRMS) and nuclear magnetic resonance spectroscopy[33]. Metabolomic techniques are increasingly supported as an approach to the discovery of biomarkers in cancer, because they explore an intermediate stage between genotype and phenotype[34]. PC is characterized by a nutrition-deficient and hypoxic condition in which dense connective tissue and poor angiogenesis predominate[35]. In order to adapt to these conditions, PC cells reprogram their metabolism to enable their survival and the maintenance of elevated proliferation rates, through the so-called Warburg effect[36]. These changes lead to the presence of specific metabolites in the serum of patients with PC, and these can serve as biomarkers.
A wide range of studies have proposed the usefulness of metabolomics, in a targeted or untargeted approach, for the discovery of novel biomarkers for PC. Our research group recently demonstrated the potential of untargeted metabolomics by liquid chromatography coupled to HRMS as a diagnostic tool for PC. We proposed a multivariate model based on nine metabolites that discriminated between PC patients and healthy controls with an area under the ROC curve (AUC) of 0.992; all 60 healthy control samples were correctly classified, and only 4 of the 59 PC samples were misclassified[37].
In a similar study, comparing between 60 patients with PC and 60 healthy controls, Luo et al[38] proposed a panel of five differentially-expressed metabolites for the diagnosis of PC (β-sitosterol, sphinganine, creatine, inosine, and glycocholic acid). In comparison with the conventional biomarkers CA 125, CA 19-9, CA 242, and CEA, their panel proved more accurate and specific to diagnose PC. The same authors also proposed two more plasma biomarkers (succinic acid and gluconic acid) for the diagnosis of PC progression and metastasis. They claimed that their novel biomarker panel could diagnose PC at an early stage; however, out of the 60 PC patients in their study, 24 were already in stage III/IV.
Recently, however, Moore et al[39] were critical of studies that compare patients in different stages of PC with healthy volunteers, describing this approach as inadequate to identify useful biomarkers for screening the general population. They described metabolomics as a useful tool to differentiate PC stages, which do not follow a linear stepwise progression. They applied this method in plasma samples from patients with neuroendocrine, intraductal papillary mucinous neoplasm (IPMN), localized PC, locally advanced PC, or metastatic PC. They described various combinations of metabolites associated with distinct PC stages, thereby facilitating differentiation between early and late PC, the follow-up of patients with pancreatic lesions, and the identification, in combination with CA 19-9 Levels, of patients with advanced disease who are candidates for resection.
Geiser et al[40] also applied metabolomics in a study of the progression from pancreatic cystic neoplasm (PCN) to PC. They proposed the combination of preoperative metabolomics with perioperative lipidomic analysis in plasma and cyst fluid from patients to support the differential diagnosis of PCN. Their integrated data modeling allowed them to discriminate between IPMN and serous cystic neoplasm with 100% accuracy and to discriminate between IPMN, and invasive cancer with an accuracy of 90.06%.
In a different approach, Zhang et al[41] studied alterations that may be discriminative for PC in 550 serum samples, analyzing the preoperative and postoperative metabolome in training cohorts. In combination with the clinical data of the patient, three discriminative metabolites (docosahexaenoic acid, lysoPC (14 : 0), and histidinyl-lysine) proved to be independent predictors of PC diagnosis. They found that the concentrations of these metabolites in postoperative patients were closer to those in healthy controls than in the paired preoperative PC group, and they obtained > 97% accuracy for this panel to differentiate between PC patients and healthy controls in an independent validation analysis.
Mayerle et al[42] undertook one of the largest metabolomic investigations in PC in order to obtain a metabolic profile that allowed PC to be distinguished from CP. In three consecutive studies, they enrolled 914 subjects in four groups (PC, CP, liver cirrhosis, healthy, and non-pancreatic disease) and were able to identify a biomarker signature composed of nine metabolites and CA 19-9. The authors stated that the clinical use of this panel would have improved the diagnosis and treatment stratification of PC in one-third of their patients in comparison to CA 19-9 alone, achieving an AUC of 0.94, with a sensitivity of 89.9% and specificity of 91.3% to distinguish between PC and CP.
Another study of major clinical interest demonstrated the value of serum metabolites in the differential diagnosis between PC and distal cholangiocarcinoma, which share many symptoms, are not easily distinguishable by imaging techniques, and require distinct management approaches. In this study, Macias et al[43] developed a logistic regression model based on a panel of nine metabolites and CA 19-9 that achieved an AUC value of 0.89 to discriminate between patients with these diseases.
The serum metabolome has also proven useful to predict recurrence in patients with resected PC. Rho et al[44] studied the preoperative serum metabolome of 57 patients undergoing pancreatectomy and developed a nomogram based on CA 19-9 alongside three metabolites, which showed a Harrell's concordance index of 0.823 and an AUC of 0.816 to predict 6 mo and 1-year cancer recurrence-free survival after surgery.
In summary, because the metabolome is the closest level to the phenotype, metabolomics is attracting increasing research interest as a promising technology to discover potential biomarkers for cancer diagnosis and for monitoring the metastatic status and therapeutic response of patients. However, most metabolomic studies have not been on a sufficiently large scale to confirm their clinical value[45]. Besides, a wide variety of analytical and clinical methodologies have been applied in metabolomic studies, limiting the reliability of their results. Efforts are currently focused on creating a database to gather data and protocols from these studies, including different biological samples and using different analytic techniques and various platforms[34].
Circulating tumor cells (CTCs) are released by a tumor into the blood before it is detectable. The study of CTCs has major potential for the diagnosis and prognosis of cancer in its earliest stages[46], and their levels have also been related to tumor aggressiveness and metastasization[47]. However, despite the clinical relevance of CTCs and abundant research on their role in other types of tumor, fewer data are available on their relation to PC[48].
Hugenschmidt et al[49] evaluated 242 preoperative samples from resectable PC patients and detected CTCs in 6.8% of high-risk patients and in 6.2% of patients with advanced disease, observing no CTCs in low-risk patients or in those with benign disease. The multivariate analysis showed that CTC status remained an independent prognostic factor after controlling for histological type, nodal status, and vascular infiltration. A worse outcome was observed in preoperative CTC-positive patients despite a successful tumor resection, and the authors proposed this methodology to guide treatment decisions in patients with PC.
Court et al[50] investigated the usefulness of CTCs as a preoperative biomarker to identify patients at high risk of occult metastatic PC. They identified CTCs in 78 of the 100 patients with PC but in none of the 26 with benign disease. The CTC count was also found to be correlated with higher PC stage and was described as a promising prognostic biomarker. In a prospective study of the capacity of CTCs to predict metastatic spread and survival, Effenberger et al[51] observed CTCs in 33.3% of a sample of 69 patients with PC, and reported significant progression-free survival (PFS) and OS in CTC-positive patients. The PFS was also significantly reduced in CTC-positive patients receiving chemotherapy, and CTC status was found to influence the outcome of patients with PC independently of other risk factors.
Further research is required not only to develop novel and more reliable technologies for CTC detection but also to identify new cellular markers for the accurate isolation of CTCs in patients with PC. Varillas et al[52] proposed a new microfluidic platform for the reliable isolation of CTCs and cancer stem cells (CSCs) in samples from these patients. It was tested in 24 patients with metastatic PC undergoing treatment, collecting 78 blood samples at different time points and finding that 84.4% were positive for CTCs and 70.8% for CSCs. The authors described the complementary assessment of CTCs and CSCs as useful to evaluate the response to treatment and guide therapeutic modifications.
Cell-surface vimentin was recently proposed by Wei et al[53] as a biomarker to isolate CTCs in patients with PC. They found that vimentin was highly expressed on the surface of mesenchymal-phenotype PC cells and detected both vimentin and CTCs in 76% of the patients. The authors obtained an AUC of 0.97 for the diagnosis of PC by combining vimentin and CTCs with CA 19–9. Vimentin + CTC counts were correlated with the change in tumor burden in patients undergoing resection, and significantly reduced CTC counts were observed after chemotherapy in responders to this treatment. Finally, higher CTC counts were correlated with a reduced recurrence-free survival period.
In conclusion, the study of CTCs appears to be a promising approach to the clinical management of PC, but a number of challenges remain. Relatively few studies have been performed, and most of them are at an experimental stage, using varied methodologies and small cohorts of patients. There is a need to develop a consensus on methodologies and protocols and to study larger cohorts in order to confirm the usefulness of CTCs as diagnostic markers for PC and as indicators of the prognosis and response to treatment, for which better results have been obtained[54].
Cell-free nucleic acids (cfNA) are nucleic acids (nuclear DNA, mitochondrial DNA, and various types of RNA molecule) present in biological fluids and independent of cells[55]. The cellular source of tumor-derived circulating nucleic acids remains controversial. Although most cell-free DNA (cfDNA) are derived from normal cells of the hematopoietic lineage[56], a small proportion has been related to tumors (primary or metastatic tumors or CTCs undergoing necrosis or apoptosis) and is designated circulating tumor DNA (ctDNA)[57]. Higher rates of necrosis and apoptosis during tumorigenesis have been associated with increased cfDNA concentrations. cfDNA concentrations reach a maximum of 100 ng/mL of blood in healthy individuals, but can be 4- fold to 40-fold higher in cancer patients[58]. cfDNA fragmentation follows a specific pattern of nucleosome distribution characteristic of caspase-dependent cleavage during apoptosis. Longer DNA fragments are considered to result from necrotic cell death[59]. However, there are subtle variations in the size of cfDNA depending on the method of isolation and on the tumor type and stage[60]. In this way, cfDNA fragment size was reported to have prognostic value for patients with advanced PC, with an association between shorter cfDNA fragment size and worse OS[61]. cfDNA can also be released via NETosis, a unique form of cell death. Upon activation, neutrophils play a role in defense mechanisms by inducing phagocytosis and degranulation and releasing neutrophil extracellular traps (NETs) in response to inflammation, infection, or hypoxia. NETs are web-like networks of decondensed nuclear DNA with histones, granule proteins (e.g., myeloperoxidase), and antimicrobial peptides[62]. The contribution of NETosis to the pool of cfDNA in PC is yet to be explored and may have therapeutic potential. This because PC is associated with one of the highest rates of venous thromboembolism (VTE) among cancers, significantly increasing the mortality[63], and neutrophils contribute to VTE in part by releasing NETs[64]. Administration of DNAse I to degrade cfDNA (including NETs) was found to reduce venous thrombosis in tumor-bearing mice[65].
The presence of cfNA in human blood was first described by Mandel and Métais in 1940[66]. Since then, researchers have compared mutations in tissue with modifications in soluble cfDNA. Mutant K-RAS has been studied in depth as one of the key driver genes in PC. A longitudinal study of PC patients undergoing surgery/chemo
ctDNA not only contains the same mutations as tumor cells but also shows the same epigenetic patterns (DNA methylation, histone modification, and chromatin remodeling), which have been associated with gene expression and tumor phenotypes[69]. One of the most widely studied epigenetic modifications is DNA methylation, i.e., the covalent addition of a methyl group to cytosines by DNA methyltransferases, generating 5-methylcytosine (5mC). Methylation commonly translates into repression of gene transcription. Conversely, oxidation of 5mC by ten-eleven translocation protein produces DNA demethylation (5hmC), which is generally associated with transcriptional activation[70]. Hence, changes in DNA methylation patterns play an important role in cancer, leading to the silencing of tumor suppressor genes or activation of oncogenes. Patients with PC have higher levels of hypermethylated genes in their cfDNA. A method based on methyl-CpG(cytosine nucleotide followed by a guanine nucleotide) binding (MBD) protein coupled with digital PCR was recently used to develop a panel of five DNA methylation markers (ADAMTS2, HOXA1, PCDH10, SEMA5A, and SPSB4) identified in fine-needle aspiration samples and paired blood samples. PC patients with methylation markers and/or KRAS mutation in cfDNA had a higher frequency of liver metastases, indicating a worse prognosis[71]. Methylated DNA immunoprecipitation coupled with high-throughput sequencing (MeDIP-seq) has been applied as an alternative method for the genome-wide detection of cfDNA methylation profiling. Li et al[72] used this technique to compare differentially methylated regions of cfDNA in PC patients and healthy controls and constructed a diagnostic prediction model that included eight differentially methylated markers (MAPT, SIX3, MIR663, EPB41L3, FAM150A, TRIM73, LOC100128977, and LOC100130148) for potential application in the non-invasive diagnosis of PC.
Global DNA hypomethylation and a reduction in 5hmC levels are also frequently observed in cancer. A different hydroxymethyl profile was recently described in PC patients in comparison to cancer-free individuals. Genes with the most altered hydroxymethylated profile were previously implicated in pancreas development (GATA4, GATA6, PROX1, ONECUT1, and MEIS2) or PC (YAP1, TEAD, PROX1, ONE
The utilization of epigenetic modifications has advantages over techniques based on genetic differences. The cfDNA sequence is identical to that of the genomic DNA, making it impossible to identify the tissue of origin. In contrast, the DNA methylation profile is highly tissue-specific, allowing organ-specific disease to be monitored. In addition, epigenetic modifications show a greater consistency in cancer in comparison to genetic changes[69].
In summary, cfDNA can be detected at relatively early stages of tumor development and is therefore useful for the early diagnosis of cancer, especially in occult organs such as the pancreas[76]. Given the short lifespan of cfDNA, ctDNA can be considered a more applicable, sensitive, and specific biomarker not only to diagnose cancer but also to monitor the tumor and response to therapy. Detection of ctDNA variants before and after anti-cancer therapy can yield information about the effectiveness of treatments and about tumor dynamics during treatment, facilitating the design of individualized treatments[77]. One cfDNA-based liquid biopsy test has been approved by the U.S. Food and Drug Administration to detect epidermal growth factor receptor (EGFR) mutations in the ctDNA of patients with non-small cell lung cancer who are candidates for targeted therapy with erlotinib and osimertinib[78]. Sixteen liquid biopsy-based clinical trials on diagnostic biomarkers for PC are under way, combining cfDNA analyses in blood and other body fluids such as intra-cystic fluid.
Another cfNA subset, composed of circulating microRNAs (miRNA), has recently become a new approach to liquid biopsy. miRNAs are short (19–24 nucleotides in length) non-coding RNAs that regulate messenger RNA (mRNA) or protein levels by promoting mRNA degradation or decreasing protein translation through an improper binding at 3-untranslated region. miRNAs can be synthesized by virtually all of the cells in the body. Many of them are ubiquitously expressed but others are tissue-specific, depending on the transcriptional and post-transcriptional regulation of miRNA precursors within the cell[79]. miRNAs can be released into the extracellular circulation and blood by passive or active secretion. Some miRNAs have been found packaged in exosomes derived from multivesicular bodies (see next section). Others may be exported in the presence of RNA-binding proteins, which give stability to the RNA molecule and make it more resistant to RNAse-mediated degradation. This stability represents a major advantage for the handling and processing of samples, because miRNA can resist degradation at room temperature for up to 4 d, during boiling and multiple freeze-thaw cycles, and at high or low pH values. Whether the miRNA is predominantly exosomal or vesicle-free depends on the miRNA itself, the cell type from which it originates, and/or other factors affecting miRNA secretion in individuals[80].
Interestingly, miRNAs of body fluids tend to be positively correlated with various human tissues[81]. Expression patterns of miRNAs (miRNA signatures) are unique to individual tissues and differ between cancer and normal tissues. Some miRNAs are overexpressed or downregulated exclusively or preferentially in certain cancer types. The fact that miRNAs are present in various body fluids, are stable, and may reflect the pathophysiologic condition of the tissue of origin has attracted interest in them as a promising group of biomarkers. It has been reported that miRNAs can be detected in the serum of cancer patients decades before clinical manifestation of their malignancy[82]. Besides serving as cancer biomarkers, tumor-derived miRNAs can act as intercellular signaling molecules modulating non-tumor cells to the benefit of the tumor[83]. miRNAs play a major role in carcinogenesis as either tumor suppressor miRNAs or oncogenic miRNAs (also termed oncomiRs). Deregulation of any miRNA type contributes to the development of tumors. Several miRNAs are known to be tumor suppressors, and their downregulation is implicated in the initiation and progression of PC. Wang et al[84] selected a panel of four miRNAs known to be overexpressed in PCs (miR-21, miR-210, miR-155, and miR-196a) in order to evaluate their usefulness as blood-based biomarkers of the disease. They found that miR-21 inhibition reduces proliferation, migration, and invasion and delays the progression of pancreatic intraepithelial neoplasia (PanIN) to PC in mouse models[85].
Vila-Navarro et al[86] very recently identified a panel of 14 circulating miRNAs (let-7e-5p, let-7f-5p, miR-103a-3p, miR-151a-5p, miR-151b, miR-16-5p, miR-181a-5p, miR-192-5p, miR-21-5p, miR-221-3p, miR-23a-3p, miR-320a, miR-33a-3p, and miR-93-5p) that are significantly overexpressed in the plasma of patients with PC and IPMN in comparison to healthy individuals. They described them as potential noninvasive biomarkers to indicate a neoplastic process in the pancreas. Two miRNAs, miR-181b-5p and miR-548d-3p, were significantly upregulated in PC plasma but not in pre
Mazza et al[89] studied three miRNAs overexpressed in patients with PC (miR-122-5p, miR-1273g-3p, and miR-6126). They obtained greater power to distinguish PC patients from healthy individuals by using plasma miR-1273g-3p levels in combination with CA 19-9 than by considering CA 19-9 alone, achieving a gain in sensitivity and negative predictive value and, therefore, a low false-negative rate. A role was proposed for these miRNAs in predicting the clinical outcomes of PC patients. Thus, higher miR-1273g-3p levels were associated with a more advanced tumor stage, and increased miR-122-5p expression emerged as an independent negative prognostic factor. In another study, the expression of miR-181b, miR-196a, and miR-210 was significantly upregulated in PC patients, and the diagnostic value for PC of each individual miRNA was increased when combined with CA19-9[90]. This panel was also associated with the prognosis, given that the expression of miR-181b, miR-196a, and miR-210 was correlated with lymph node metastasis, clinical stage, and vascular invasion. In another study, a panel of miR-99a-5p, miR-200c-3p, and miR-365a-3p proved able to discriminate PC patients with a poor post-resection outcome from those with longer survival[91].
Gemcitabine resistance has also been associated with a modulation in miRNA expression. MiR-125a-3p is a tumor suppressor found to be downregulated in plasma samples from patients with PC[92]. In vitro studies showed that miR-125a-3p can directly inhibit the expression of Fyn, thereby promoting the epithelial-to-mesenchymal process, and that Fyn overexpression can partially reverse miR-125a-3p-mediated chemoresistance to gemcitabine[93]. Numerous miRNAs have been associated with resistance to chemotherapy; however, most results have been obtained in in vitro studies, and the potential role of these miRNAs as blood-based prognostic biomarkers has yet to be validated.
As stated above, PC cannot be considered as a single disease but rather a combination of different subtypes. Kandimalla et al[94] identified a panel of nine miRNAs that were significantly upregulated (miR-205-5p and -934) or downregulated (miR-192-5p, 194-5p, 194-3p, 215-5p, 375-3p, 552-3p, and 1251-5p) in PC molecular (squamous and quasi-mesenchymal) subtypes associated with poor survival outcomes. They first analyzed the International Cancer Genome Consortium dataset and The Cancer Genome Atlas dataset to identify nine tissue miRNAs specifically deregulated in both PC subtypes and observed that six of them had similar expression profiles in serum specimens. The five-year OS rate was higher in PC patients with high-risk scores for these miRNAs in serum than in those with low-risk scores. These findings confirm the usefulness of this miRNA profile to identify poor molecular subtypes and high-risk PC patients and to predict their prognosis.
It was also demonstrated that a panel of seven miRNAs (miR-486-5p, miR-106b-5p, let-7i-5p, let-7g-5p, miR-144-3p, miR-19a-3p, and miR-103a-3p) could estimate the risk of future VTE in patients with PC at their diagnosis. These miRNAs are implicated in the PC pathway and in complement and coagulation cascades[95].
Bioinformatic tools allow the integration of different PC-related serum miRNA expression profiles to determine the most relevant miRNA signatures. In this way, Shams et al[92] constructed a novel miRNA-mRNA regulatory network for PC. They proposed a panel of downregulated miRNAs (miR-125b-1-3p, miR-125a-3p, miR-92a-5p, miR-4530, miR-6893-5p, and miR-4476) and found that a combination of miR-125a-3p, miR-92a-5p, and miR-4530 was the most promising panel to differentiate PC patients from healthy controls, with an AUC of 0.95, sensitivity of 0.98, and specificity of 0.97. Among other upregulated miRNAs (miR-1469, miR-1246, miR-5100, miR-8073, miR-642b-3p, and miR-663a), miR-1469 was found to be the most powerful individual marker for PC diagnosis.
Five of the aforementioned genes (miR-125a-3p, miR-6893-5p, miR-125b-1-3p, miR-1469, and miR-4476) were also selected by Yan el at[96] for inclusion in a model with another eight miRNAs (miR-6075, miR-6836-3, miR-6729-5, miR-575, miR-204-3, miR-6820-5, miR-4294, and miR-4792), and this 13-miRNA panel accurately differentiated between PC patients and healthy individuals. However, none of these models have been clinically validated, and further research is needed before their application can be recommended.
Most miRNAs that have been identified as cancer biomarkers have failed to advance from the experimental stage to a clinical trial. This is largely attributable to the wide variability of results and the lack of reproducibility. Further standardization and technological improvements are needed before miRNAs can be used to obtain an accurate and reliable cancer diagnosis in the clinical setting[97]. In this regard, elucidation of the miRNA expression profile of individual tumors could improve diagnostic accuracy, treatment selection, and prognosis prediction.
Virtually all types of cells are able to release phospholipid bilayer-enclosed extracellular vesicles (EVs). Their diameter ranges from 30 nm to 5000 nm, and they are mostly classified as exosomes (30–120 nm in diameter), microvesicles (MVs, also known as ectosomes, or microparticles, 100–1000 nm), or apoptotic bodies (ranging from 800 to 5000 nm)[98]. Beyond their role in the elimination of cellular waste, EVs are potent vehicles of cell-to-cell communication. They transport all types of biomolecule (proteins, lipids, and nucleic acids) under both physiological and pathological conditions. EVs can use their specific cargo to modulate the physiology of recipient cells, including processes that are deregulated in human cancers (e.g., proliferation, angiogenesis, and apoptosis)[99]. Encapsulation of the cargo in a membrane gives it a higher stability, longer half-life, greater resistance to degradation, and an increased capacity to travel long distances in comparison to free proteins, lipids, and nucleic acids in the cytoplasm. Moreover, this specific cargo provides a rich source of biomarkers that can be found in all body fluids[100] . EVs are formed by distinct mechanisms. MV biogenesis occurs through direct outward blebbing and pinching of the plasma membrane, releasing the nascent MV into the extracellular space. Exosomes originate from inward blebbing and budding (invagination) of the plasma membrane to form the early endosome that forms the late endosome after a series of modifications, and is secreted upon fusion with the cell surface[101]. Ultimately, the different particles are differentiated by their size, biogenesis, surface markers, and cargo content.
PC is characterized by a complex microenvironment with constant interaction between PC cells and stromal cells such as fibroblasts, myofibroblasts, stellate cells, vascular endothelial cells, and immune cells, which are all surrounded by an abundant fibrotic extracellular matrix that serves as a shield[102]. These interactions are decisive steps in tumor progression and are at least in part mediated by EVs[103], giving exosomes a key role in PC.
The cargos of exosomes largely comprise proteins, RNAs, and lipids (ExoCarta-Exosome dataset), and these molecules can all be used as diagnostic, predictive, and/or prognostic markers for PC. Table 1 summaries the latest panels described in the literature regarding exosome’s cargo[104-113]. The fact that almost all cells can release exosomes into the peripheral circulation is one of the major drawbacks of their utilization as tumor surrogates. The dilution of tumor exosomes with non–cancer cell-derived exosomes means that blood samples must be enriched in tumor-specific exosomes. Some of the surface proteins described in the exosomes have been used for that purpose[114]. One of the most studied components of the exosomes are the miRNA, but there are other types of RNA molecules that have been explored as biomarkers for PC. Interestingly, while extracellular circulating levels of some miRNA did not differ when comparing PC patients and healthy individuals, exo-miRNAs did. Consequently, some researchers have pointed to miRNA encapsulated in exosomes as a more useful source of biomarkers[115]. Although other biofluids are sources of exosomes, authors advised against evaluating pancreatic juice from patients with IPMN, because exosomes could not be reliably extracted due to the high viscosity of the mucin content[116]. Although most of the studies have focused on exo-miRNAs profiling, whole transcriptome analysis has identified novel disease biomarkers. Besides, very recently circular RNA (circRNA) has been recently recognized as a novel class of highly stable noncoding RNA species that is abundant in exosomes and that are generated from the ligation of exons, introns, or both. CircRNAs can function by binding to miRNAs as sponges and suppress miRNA activity. As miRNAs are known to alter the development and progression of cancer, circRNAs may offer a novel diagnostic and prognostic biomarker for cancer[117]. Most of the research carried out analyzing circRNA have been performed in tissue samples[118] and their potential as blood-based biomarkers for PC is unexplored.
| Proteins | Source | Outcome | Utility | Ref. |
| CLDN4, EPCAM, CD151, LGALS3BP, HIST2H2BE, and HIST2H2BF | Blood | Higher KRAS mutation in exosomes | Protein panel PC exosome-enriched samples | Castillo et al[114] |
| TAAs | Plasma | Higher levels of circulating Ig-bound exosomes in PC patients than in healthy individuals | Circulating AAbs against TAAs to discern between PC, CHP and healthy individuals | Capello et al[104] |
| CEACAM 1/5 and tenascin C | Pancreatic duct fluid | Upregulation in PC | Distinguish patients with PC/IPMN or CHP | Zheng et al[105] |
| RNA | ||||
| exo-miR155 | Plasma Tissue | GEM-resistant cells secreted higher levels of exo-miR-155 miR-155 expression level induced exosome secretion | Predict resistance to Gemcitabine. Novel therapeutic target | Mikamori et al[106] |
| exo-miR-10b, miR-21, miR-30c, miR-181a and miR-let7a | Serum | Upregulation in exo-miR-10b, miR-21, miR-30c, miR-181a; Repression in miR-let7a | Distinguish patients with CP/CHP or other pancreatic disease; PC resection monitoring | Lai et al[107] |
| exo-miR-451a | Serum | Upregulation in stage II PC with recurrence after surgery | Biomarkerofrecurrence | Takahasi et al[108] |
| exo-miR-191, miR-21 and miR-451 | Serum | Dysregulated in IPMN and PC patients | Diagnosticbiomarkers | Goto et al[115] |
| exo-miR-21 and exo-miR-155 | Pancreatic juice | Upregulation in PC patients compared with CHP | Diagnostic of early-stage PC | Nakamura et al[116] |
| miR-4525, miR-451a, and miR-21 | PVB | Upregulation of the panel | Identify patients at high risk for recurrence and poor survival after PC resection | Kawamura et al[109] |
| mRNA | ||||
| MMP8, TBX3, PDX1, CTSL, SIGLEC15, IL32, SIGLEC11, DCN, HOXA5, KLRB1 | Serum | MMP8, TBX3, PDX1, CTSL, SIGLEC15 overexpression in PC/IPMN vs healthy; IL32, SIGLEC11, DCN repression in PC vs IPMN and healthy; HOXA5, KLRB1 repression in PC/IPMN vs healthy | Identify different subtypes | Kumar et al[110] |
| CCDC88A, ARF6, VAV3, and WASF2 | Serum | Upregulation between PC and control patients | PC risk and PC diagnosis | Kitagawa et al[111] |
| circRNA | ||||
| circPDE8A; circIARS | Tumor tissues Plasma | Upregulation in PC | Predictlivermetastasis, vascular invasion, and tumor-node-metastasis (TNM) stage and por postoperativesurvival time | Li et al[112] |
| circPC | Tumor tissues Serum | Upregulation in PC | Diagnostic biomarker | Seimiya et al[113] |
Based on their role as biomolecule carriers and because they are non-immunogenic particles, exosomes have been proposed as new disease therapy strategies not without complications and challenges mainly regarding regulatory production guidelines, production of sufficient number of safe, high quality and efficient exosomes. A better understanding of the molecular and cellular processes regulating exosome biogenesis is expected to increase technological advances and potential clinical applications[119,120]. Furthermore, the pathways on which any therapeutic miRNA functions would need to be extensively defined, to avoid undesirable off-target effects[121]. A major drawback is that currently there is no state-of-the-art technology to isolate EVs, for either therapeutic application or basic research which can impose lack of reproducibility. Exosome-based therapies are been tested. For PC, a phase I study is being developed by the University of Texas M.D. Anderson Cancer Centre in Houston, Texas (NCT03608631) to deliver KrasG12D siRNA (given the drug loading capacity of exosomes) to treat metastatic pancreas cancer with KrasG12D mutation. Patients will receive the exosomes intravenously and outcomes will be evaluated over time[119].
Cytokines participate in intercellular communications and are highly relevant to tumor growth and metastasization[122,123]. They modulate cells, the extracellular matrix, and immune system communications in patients with PC[124-126]. In addition, the overexpression of growth factors such as insulin-like growth factor (IGF), epidermal growth factor (EGF), transforming growth factor-alpha and beta (TGFα and TGFβ), fibroblast growth factor (FGF), and an elevated expression of their cancer cell surface receptors have been reported in many gastrointestinal cancers, including PC. All of these factors trigger an intracellular signaling cascade that leads to cell proliferation, invasion, and survival. Overexpression of these proliferation factors is generally associated with a poor prognosis[125,127].
Another factor that influences the aggressiveness[125] and resistance to treatment[128-130]of PC is the specific microenvironment of the tumor, where its inflammatory nature is reflected. The PC microenvironment is characterized by hypoxia and an increased desmoplasia[131,132]. The stromal network comprises a large amount of extracellular matrix produced by pancreatic stellate cells (PSCs) and cancer-associated fibroblasts (CAFs). Various types of cell can be found in this matrix, including endothelial cells, tumor-associated macrophages, Th cells, myeloid-derived suppressor cells, dendritic cells, and natural killer cells[133]. These can all release pro-inflammatory cytokines, growth factors, and other local mediators that increase the aggressiveness of the tumor[134], as well as immunosuppressive cytokines[135]. Taken together, these factors and the physical barrier of the stromal network can prevent penetration of PCs by the immune system or treatments[129].
Growth factors reported to regulate desmoplasia and cellular growth in PC include EGF, IGF, FGF, TGFα, TGFβ, and hepatocyte growth factor (HGF)[136]. Overexpression of EGFR has been described in 30%–95% of PC cases and is associated with a poor prognosis[137]. Its overexpression is moderate in CP and is not detected in healthy peritumoral tissue, suggesting its involvement in the malignant transform
IGF and IGF receptor are expressed on pancreatic cells, and their activation has been implicated in cell proliferation, invasion, and survival. The binding proteins (IGFBPs) that can inhibit IGF-1 have also attracted research interest as possible prognostic markers for numerous diseases, including PC[138,139]. IGFBP2 overexpression was observed in PanIN lesions, suggesting its possible usefulness as a marker of early disease[140].
TGF is a key regulator of cell growth and apoptosis in PC[141] , and alterations in its signaling have been associated with early and advanced stages of gastrointestinal cancer, especially in PC[142]. Elevated TGFβ was not detected in pancreatic lesions by some authors[143], whereas TGFβ-1, TGFβ-2, and TGFβ-3 isoforms were correlated with more advanced cancer PC stage and worse survival outcomes[144].
No association has been found between FGF and PC development, but the FGF receptor (FGF-R) has been correlated with very advanced stages when the tumor size is large. Consequently, the overexpression of FGF-R may be a marker of short-term survival[145].
The HGF/c-MET signaling pathway participates in desmoplasia regulation because it is involved in communications among PSCs[144]. It has been reported that the HGF/c-MET signaling pathway is upregulated in PC. The main producers of HGF are CAFs, promoting the malignant transformation of pancreatic cells[146].
Evidence has recently emerged on the diagnostic potential of pS2 peptide, which belongs to a newly described family of trefoil-shaped growth factors. Its expression was observed in more than 50% of PC cases but was not detected in CP[147].
As previously noted, the relationship of PC with inflammatory processes is well documented[148], and inflammation-associated conditions such as obesity or diabetes are major risk factors for PC[149]. Cytokines are the main inflammatory mediators, and some have been associated with a reduced OS in patients with PC, including interleukin (IL)-6, IL-8, macrophage inhibitory cytokine-1, oncostatin M (OSM), IL-10, IL-11, leukemia inhibitory factor, cardiotrophin-like cytokine, ciliary neurotrophic factor, cardiotrophin-1, neuropoietin, IL-27, and IL-3[150,151]. Serum IL-6 Levels are increased in PC patients and are even higher in those with advanced metastatic PC[150]. In general, elevated cytokine expression has been correlated with cachexia, performance status, and survival in patients with PC[151]. The immunosuppressive cytokine IL-10 has also been implicated in immune system evasion in cases of PC[152].
Although each individual cytokine is of limited value as a biomarker, panels of cytokines have proven useful to predict a poor prognosis in PC, including the combination of B7-1/CD80, EG-VEGF/PK1, IL-29, NRG1-beta1/HRG1-beta1, and PD-ECGF, which may therefore represent novel therapeutic targets for this disease[153]. A panel composed of FGF-10/KGF-2, I-TAC/CXCL11, OSM, osteoactivin/glycoprotein nonmetastatic melanoma protein B, and SCF has also been proposed as a diagnostic biomarker[154].
Responses of the immune system to cancer, especially the humoral response, have been widely investigated[155,156], and the immune system is known to generate autoantibodies (AAbs) against tumor antigens (TA)[157]. Tumor cells produce proteins altered by mutations, misfolding, or post-translational modifications such as glycosylation or simple overexpression. The immune system produces AAbs against these new TAs, although the underlying mechanism of action has not been fully elucidated[158]. Considerable research has addressed the question why some patients develop a humoral response against TAs but others do not[159].
AAb production is an early event in tumor development, enhancing their potential value as tumor markers. In addition, they survive for a long time period without degradation and in a higher concentration than antigens TAs, with an amplified signal[160]. The “cancer seroma” has been described as the repertoire of antigens recognized by the humoral immune system[161], and research efforts have focused on the search for AAb or antibody signatures with predictive value as cancer biomarkers when considered alone or in combination with others[160].
As many as 124 AAbs have been associated with PC but often in only one study with no replication, and 86% of them offered a sensitivity lower than 50%[158]. Four of these AAbs met sensitivity and specificity criteria: anti-CLP peptide, anti-Mesothelin, anti-Ezrin, and anti-ENOA1,2[162,163].
The low prevalence of PC means that biomarkers should be highly specific[164]. It is difficult to properly correlate AAbs with PC for diagnostic purposes because of the low immune response to this cancer, attributable to the protection offered by the stromal network and immunosuppressive mechanisms[159-161]. Additional research efforts are therefore warranted to systematize the study of tumor stage and to improve detection methodologies[159].
Other AAbs have been studied, including MUC1[165], p53, calreticulin isoform 1 or 2, and Rad51[157]; however, their detection is not always linked to a poor prognosis, as in the case of AAbs against MUC-1[165]. Moreover, they are not only detected in PC but also in other types of tumor (e.g., MUC-1, mesothelin, ezrin, ENOA) or in other pancreatic diseases such as CP (e.g., p53, ENO, or ANX), resulting in a low specificity[158].
Some AAbs have been related to tumor stage[162], showing higher sensitivity for late stages[158]. For instance, EZRIN, detected in mouse models of PanIN lesions, only meets sensitivity criteria at advanced stages of cancer in humans, although it has been suggested as an early marker[162]. Other AAbs associated with early lesions include VCL, PDC&I, hnRNPL, VIM, K2C8, or ANXA[166].
A further concern is the low capacity of AAbs to differentiate between PC and other inflammatory diseases, especially CP or autoimmune pancreatitis, which are responsible for a large production of AAbs. In this regard, some authors have proposed a panel of PP1R15A, CYP3A5, and WDR45 antigens to discriminate between PC and benign inflammatory disease[164].
Some researchers have suggested that AAbs do not yield reliable information on the localization of cancers and may best serve to screen parents for the presence of cancer in general, followed by more specific diagnostic measures when a positive result is obtained[158].
Platelets are enucleated cells released as cytoplasmic bodies from megakaryocytes in bone and lung[167]. They have attracted research interest for the diagnosis and follow-up of tumors[168,169] due to the biochemical and physiological alterations undergone during disease[170]. The short half-life (5-10d) of platelets represents a further advantage, allowing the disease to be monitored in almost real time[171].
It has been reported that interaction between platelets and cancer produces platelet hyperactivity, increasing the number of young platelets in cancer patients and reducing their count during post-treatment remission[172]. Cancer patients are known to be in a state of hypercoagulation and to have altered platelets, increasing the risk of thromboembolic disease. It has been proposed that metastasis, immune evasion, and/or angiogenesis in the tumor environment are favored by relationships among the blood coagulation system, platelet functions, and the spread of cancer through the blood[173].
VTE has been observed in around 60% of patients with PC[173], in part related to tissue factor (TF) expression[174]. TF is expressed during malignant transformation to PC, showing four-fold higher levels in cancer, endothelial, inflammatory, and stromal cells[174]. EVs produced by cancer cells to communicate with endothelial cells also exert pro-thrombotic effects, indirectly inducing the over-activation of platelets and the hypercoagulability state that characterizes PC[175]. Platelets are also activated through interaction with mucins released into the circulation by the tumor[176]. Inhibition of fibrinolysis has also been described, and all of these factors can produce a systemic hypercoagulable state in these patients[177].
A direct relationship has been established between tumor stage and platelet count. Chen et al[178] recently described for the first time a relationship between platelet count and the post-treatment survival of patients with PC, correlating the platelet count with the concentration of CA19-9. The prognostic value of the pre-treatment platelet/Lymphocyte ratio (PLR) has also been studied in PC, but the results have not been conclusive[179]. Although some authors related this ratio to a poor prognosis, more recent studies associated a high PLR with a better OS rate[179,180].
The activation of platelets increases their size, and the mean platelet volume (MPV) has been studied as an indicator of their activation[181]. Changes in MPV have been detected in various types of cancer[182], and its increase has been associated with worse survival and the development of liver metastases in patients with PC[183].
In a process of mutual communication, tumor cells transfer biomolecules to platelets, generating so-called tumor-educated platelets (TEPs)[169,170]that have been implicated in the progression and spread of solid tumors. Resulting alterations in the composition of platelets generate a dynamic flow of biomolecule exchange in the tumor microenvironment that can be used as a tumor biomarker[170,184,185]. Platelets physiologically interchange RNA and proteins with other platelets, immune system cells, endothelial cells, and tumor cells[171]. The composition of platelets can change in two ways. On one hand, interactions of platelets with the tumor environment produce changes in RNA maturation (mRNA and miRNA from platelets) and their transduction to proteins[186]. On the other hand, their composition can be modified by the direct transfer of biomolecules through tumor MVs. In this way, miRNAs in platelet-derived MVs can regulate the gene expression of tumor cells that phagocytose them. The mechanisms underlying the bi-directional selection and internalization of vesicles remain unknown[171]. Both of the above mechanisms generate a dynamic flow of biomolecules between platelets and tumor cells that can provide complete and updated information on the tumor in both space and time, given that platelets are renewed after around one week. TEPs are therefore a promising tool for the constant monitoring of cancer, especially for cancers of difficult access such as PC.
As noted above, this interchange of biomolecules involves RNA, miRNA from platelets, and proteins, which are all potential cancer markers. First, the characteristic profiles or signature of tumor RNA can offer specific information on the presence, localization, and molecular features of a cancer[171]. For instance, mRNA onco-signatures in TEPs from PC allowed differentiation between patients with PADC and other types of cancer and even between PC patients with KRAS mutant tumor and those with KRAS wild-type tumor[169]. RNA in TEPs can derive from the tumor itself or from an alteration in RNA processing (e.g., intron retention in mRNA), resulting in RNA patterns that differ from those in normal platelets[187]. With regard to miRNA, which is highly abundant in platelets[188], isomiRs generated by failures in miRNA maturation or modification of its ends are valuable biomarkers of cancer[189]. Finally, platelets contain a large number of proteins with biological functions derived from their synthesis or from the circulation. Data on proteins in platelets from the tumor environment may therefore be useful as cancer markers[186].
In 2015, Best et al[169] were the first to use mRNA profiles of TEPs in the diagnosis of cancer. They developed two diagnostic algorithms based on TEP RNA sequencing data that were able to detect the presence of cancer with a sensitivity of 97%, specificity of 94%, and accuracy of 96%. These algorithms also obtained an overall accuracy of 71% for an organ-specific cancer diagnosis in patients with breast cancer, colorectal cancer, glioblastoma, hepatobiliary cancer, lung cancer, or PC. The detection rate of these algorithms was 50%-62% for PC and 71% for gastric cancer. With the information generated by this study, Zhang et al[190] developed an algorithm for the analysis of gene expression profiles in blood. The following 18 genes distinguished the above cancer subtypes and healthy samples: ribosomal protein S20; ribosomal protein lateral stalk subunit P2; T cell receptor beta constant 2; ribosomal protein SA; ribosomal protein S11; ribosomal protein L11; ribosomal protein S16; CD3g molecule; Ras homolog family member H; CD27 molecule; ribosomal protein L27; ribosomal protein L9; ribosomal protein L31; ATM serine/threonine kinase; FAU, ubiquitin-like and ribosomal protein S30 fusion; ribosomal protein L34; and ribosomal protein L6.
Mantini et al[186] conducted a wide study designed to integrate and compare miRNA, mRNA, and protein omics data between platelets from patients with PC and from those with benign disease. Interestingly, SPARC protein was downregulated in platelets from PC patients but was negatively correlated with miR-17-3p, miR-29a-3p, miR-22-3p, and miR-221-5p. These miRNAs are associated with the overexpression of classical pathways in PC such as ErbB, PI3K-Akt, and mTOR while SPARC is a negative regulator of proliferation, adhesion, and angiogenesis. In addition, platelets from PC patients contained numerous miRNAs related to mRNA splicing, but the amounts of mature mRNA were low. As a possible explanation, the authors proposed that these miRNAs and proteins were exogenous to the platelets and derived from the tumor.
Analysis of the contents of platelets appears to be a promising tool for the in-situ diagnosis of PC and for tracking TEPs in liquid biopsies as a potential biomarker of early-stage PC. However, as in the case of ctDNA, its utilization and accuracy must be optimized to allow determination of the cancer subtype[169].
As highlighted throughout this review, liquid biopsies offer major opportunities to improve the screening, treatment guidance, and follow-up of cancer patients in general and patients with PC in particular. This is extremely important for patients with PC because there can be a time lapse of up to 20 years between the first appearance of lesions and development of the disease, providing a wide time window for preventative interventions. Among other advantages, the liquid biopsy is a non-invasive and readily-accessible source of information, and samples can be taken as often as needed to monitor the progress of the patient. Liquid biopsies can be used to obtain an early diagnosis, to estimate the risk of relapse or metastatic progression, and to follow up the therapeutic response in real time, allowing futile treatments to be suspended or avoided. Nevertheless, this approach has some limitations, including an inadequate sensitivity due to a low concentration of biosources such as cfDNA or CTCs in liquid biopsies. A further challenge is posed by the lack of standardization and the absence of broadly accepted standard operating procedures for liquid biopsy evaluation. Different isolation methodologies have yielded sensitivity values ranging between 0.27 and 0.95, giving a pooled sensitivity of only 0.64. The final result is influenced by the matrix (blood or serum), time interval between sample collection and processing, type of tube used, centrifugation parameters, and sample handling conditions, among other factors. This may explain the enormous number of molecular signatures described by different research groups. Considerable research efforts are required to verify the diagnostic and prognostic value of liquid biopsies in PC before their routine clinical implementation can be recommended.
Manuscript source: Invited manuscript
Corresponding Author's Membership in Professional Societies: American Association for Cancer Research (AACR), No. 368150.
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Spain
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Wang XB S-Editor: Ma YJ L-Editor: A P-Editor: Li JH
| 1. | Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7-30. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12667] [Cited by in F6Publishing: 13859] [Article Influence: 3464.8] [Reference Citation Analysis (4)] |
| 2. | Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74:2913-2921. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3888] [Cited by in F6Publishing: 4587] [Article Influence: 458.7] [Reference Citation Analysis (0)] |
| 3. | Martin-Perez E, Domínguez-Muñoz JE, Botella-Romero F, Cerezo L, Matute Teresa F, Serrano T, Vera R. Multidisciplinary consensus statement on the clinical management of patients with pancreatic cancer. Clin Transl Oncol. 2020;22:1963-1975. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 21] [Article Influence: 5.3] [Reference Citation Analysis (1)] |
| 4. | Moore A, Donahue T. Pancreatic Cancer. JAMA. 2019;322:1426. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 55] [Cited by in F6Publishing: 77] [Article Influence: 15.4] [Reference Citation Analysis (1)] |
| 5. | McGuigan A, Kelly P, Turkington RC, Jones C, Coleman HG, McCain RS. Pancreatic cancer: A review of clinical diagnosis, epidemiology, treatment and outcomes. World J Gastroenterol. 2018;24:4846-4861. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 906] [Cited by in F6Publishing: 968] [Article Influence: 161.3] [Reference Citation Analysis (30)] |
| 6. | Duffy MJ. Tumor markers in clinical practice: a review focusing on common solid cancers. Med Princ Pract. 2013;22:4-11. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 184] [Cited by in F6Publishing: 145] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 7. | Le N, Sund M, Vinci A; GEMS collaborating group of Pancreas 2000. Prognostic and predictive markers in pancreatic adenocarcinoma. Dig Liver Dis. 2016;48:223-230. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 84] [Cited by in F6Publishing: 95] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 8. | Yadav DK, Bai X, Yadav RK, Singh A, Li G, Ma T, Chen W, Liang T. Liquid biopsy in pancreatic cancer: the beginning of a new era. Oncotarget. 2018;9:26900-26933. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 43] [Cited by in F6Publishing: 42] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 9. | Luchini C, Veronese N, Nottegar A, Cappelletti V, Daidone MG, Smith L, Parris C, Brosens LAA, Caruso MG, Cheng L, Wolfgang CL, Wood LD, Milella M, Salvia R, Scarpa A. Liquid Biopsy as Surrogate for Tissue for Molecular Profiling in Pancreatic Cancer: A Meta-Analysis Towards Precision Medicine. Cancers (Basel). 2019;11. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 28] [Article Influence: 5.6] [Reference Citation Analysis (1)] |
| 10. | Scarà S, Bottoni P, Scatena R. CA 19-9: Biochemical and Clinical Aspects. Adv Exp Med Biol. 2015;867:247-260. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 144] [Cited by in F6Publishing: 176] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 11. | Goh SK, Gold G, Christophi C, Muralidharan V. Serum carbohydrate antigen 19-9 in pancreatic adenocarcinoma: a mini review for surgeons. ANZ J Surg. 2017;87:987-992. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 29] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 12. | Fahrmann JF, Schmidt CM, Mao X, Irajizad E, Loftus M, Zhang J, Patel N, Vykoukal J, Dennison JB, Long JP, Do KA, Chabot JA, Kluger MD, Kastrinos F, Brais L, Babic A, Jajoo K, Lee LS, Clancy TE, Ng K, Bullock A, Genkinger J, Yip-Schneider MT, Maitra A, Wolpin BM, Hanash S. Lead-Time Trajectory of CA19-9 as an Anchor Marker for Pancreatic Cancer Early Detection. Gastroenterology. 2021;160:1373-1383.e6. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 69] [Article Influence: 23.0] [Reference Citation Analysis (1)] |
| 13. | Hegyi P, Párniczky A, Lerch MM, Sheel ARG, Rebours V, Forsmark CE, Del Chiaro M, Rosendahl J, de-Madaria E, Szücs Á, Takaori K, Yadav D, Gheorghe C, Rakonczay Z Jr, Molero X, Inui K, Masamune A, Fernandez-Del Castillo C, Shimosegawa T, Neoptolemos JP, Whitcomb DC, Sahin-Tóth M; Working Group for the International (IAP – APA – JPS – EPC) Consensus Guidelines for Chronic Pancreatitis. International Consensus Guidelines for Risk Factors in Chronic Pancreatitis. Recommendations from the working group for the international consensus guidelines for chronic pancreatitis in collaboration with the International Association of Pancreatology, the American Pancreatic Association, the Japan Pancreas Society, and European Pancreatic Club. Pancreatology. 2020;20:579-585. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 30] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 14. | Singh S, Tang SJ, Sreenarasimhaiah J, Lara LF, Siddiqui A. The clinical utility and limitations of serum carbohydrate antigen (CA19-9) as a diagnostic tool for pancreatic cancer and cholangiocarcinoma. Dig Dis Sci. 2011;56:2491-2496. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 68] [Cited by in F6Publishing: 74] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 15. | Lee T, Teng TZJ, Shelat VG. Carbohydrate antigen 19-9 - tumor marker: Past, present, and future. World J Gastrointest Surg. 2020;12:468-490. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 93] [Cited by in F6Publishing: 83] [Article Influence: 20.8] [Reference Citation Analysis (2)] |
| 16. | Parra-Robert M, Santos VM, Canis SM, Pla XF, Fradera JMA, Porto RM. Relationship Between CA 19.9 and the Lewis Phenotype: Options to Improve Diagnostic Efficiency. Anticancer Res. 2018;38:5883-5888. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 31] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 17. | Gui JC, Yan WL, Liu XD. CA19-9 and CA242 as tumor markers for the diagnosis of pancreatic cancer: a meta-analysis. Clin Exp Med. 2014;14:225-233. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 39] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 18. | Dou H, Sun G, Zhang L. CA242 as a biomarker for pancreatic cancer and other diseases. Prog Mol Biol Transl Sci. 2019;162:229-239. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 23] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 19. | Bonifácio VDB. Ovarian Cancer Biomarkers: Moving Forward in Early Detection. Adv Exp Med Biol. 2020;1219:355-363. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 57] [Cited by in F6Publishing: 49] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 20. | Wallentin L, Eriksson N, Olszowka M, Grammer TB, Hagström E, Held C, Kleber ME, Koenig W, März W, Stewart RAH, White HD, Åberg M, Siegbahn A. Plasma proteins associated with cardiovascular death in patients with chronic coronary heart disease: A retrospective study. PLoS Med. 2021;18:e1003513. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 55] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 21. | Glasgow CG, Pacheco-Rodriguez G, Steagall WK, Haughey ME, Julien-Williams PA, Stylianou MP, Gochuico BR, Moss J. CA-125 in Disease Progression and Treatment of Lymphangioleiomyomatosis. Chest. 2018;153:339-348. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 22] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 22. | Oliveira MAP, Raymundo TS, Soares LC, Pereira TRD, Demôro AVE. How to Use CA-125 More Effectively in the Diagnosis of Deep Endometriosis. Biomed Res Int. 2017;2017:9857196. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 14] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 23. | Park M, Dhawan R, Whittaker E, Kon OM. CA 125 and TB. BMJ Case Rep. 2021;14. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in F6Publishing: 3] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 24. | Gao Y, Wang J, Zhou Y, Sheng S, Qian SY, Huo X. Evaluation of Serum CEA, CA19-9, CA72-4, CA125 and Ferritin as Diagnostic Markers and Factors of Clinical Parameters for Colorectal Cancer. Sci Rep. 2018;8:2732. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 94] [Cited by in F6Publishing: 156] [Article Influence: 26.0] [Reference Citation Analysis (0)] |
| 25. | Jelski W, Mroczko B. Biochemical diagnostics of pancreatic cancer - Present and future. Clin Chim Acta. 2019;498:47-51. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 16] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 26. | van Manen L, Groen JV, Putter H, Vahrmeijer AL, Swijnenburg RJ, Bonsing BA, Mieog JSD. Elevated CEA and CA19-9 serum levels independently predict advanced pancreatic cancer at diagnosis. Biomarkers. 2020;25:186-193. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 49] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 27. | van Manen L, Groen JV, Putter H, Pichler M, Vahrmeijer AL, Bonsing BA, Mieog JSD. Stage-Specific Value of Carbohydrate Antigen 19-9 and Carcinoembryonic Antigen Serum Levels on Survival and Recurrence in Pancreatic Cancer: A Single Center Study and Meta-Analysis. Cancers (Basel). 2020;12. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 28. | Luo G, Liu C, Guo M, Cheng H, Lu Y, Jin K, Liu L, Long J, Xu J, Lu R, Ni Q, Yu X. Potential Biomarkers in Lewis Negative Patients With Pancreatic Cancer. Ann Surg. 2017;265:800-805. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 79] [Cited by in F6Publishing: 114] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 29. | Zhang Y, Yang J, Li H, Wu Y, Zhang H, Chen W. Tumor markers CA19-9, CA242 and CEA in the diagnosis of pancreatic cancer: a meta-analysis. Int J Clin Exp Med. 2015;8:11683-11691. [PubMed] [Cited in This Article: ] |
| 30. | Deng GC, Yan H, Guo ZP, Dai G. Correlation Between Baseline Serum Tumor Markers and Clinical Characteristic Factors in Patients with Advanced Pancreatic Cancer. Onco Targets Ther. 2020;13:11151-11163. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 4] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 31. | Xu HX, Li S, Wu CT, Qi ZH, Wang WQ, Jin W, Gao HL, Zhang SR, Xu JZ, Liu C, Long J, Xu J, Ni QX, Yu XJ, Liu L. Postoperative serum CA19-9, CEA and CA125 predicts the response to adjuvant chemoradiotherapy following radical resection in pancreatic adenocarcinoma. Pancreatology. 2018;18:671-677. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 32. | Khomiak A, Brunner M, Kordes M, Lindblad S, Miksch RC, Öhlund D, Regel I. Recent Discoveries of Diagnostic, Prognostic and Predictive Biomarkers for Pancreatic Cancer. Cancers (Basel). 2020;12. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 25] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 33. | Amberg A, Riefke B, Schlotterbeck G, Ross A, Senn H, Dieterle F, Keck M. NMR and MS Methods for Metabolomics. Methods Mol Biol. 2017;1641:229-258. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 42] [Cited by in F6Publishing: 46] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 34. | Cheung PK, Ma MH, Tse HF, Yeung KF, Tsang HF, Chu MKM, Kan CM, Cho WCS, Ng LBW, Chan LWC, Wong SCC. The applications of metabolomics in the molecular diagnostics of cancer. Expert Rev Mol Diagn. 2019;19:785-793. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 38] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 35. | Kamphorst JJ, Nofal M, Commisso C, Hackett SR, Lu W, Grabocka E, Vander Heiden MG, Miller G, Drebin JA, Bar-Sagi D, Thompson CB, Rabinowitz JD. Human pancreatic cancer tumors are nutrient poor and tumor cells actively scavenge extracellular protein. Cancer Res. 2015;75:544-553. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 498] [Cited by in F6Publishing: 596] [Article Influence: 66.2] [Reference Citation Analysis (0)] |
| 36. | Gu W, Tong Z. Clinical Application of Metabolomics in Pancreatic Diseases: A Mini-Review. Lab Med. 2020;51:116-121. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in F6Publishing: 4] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 37. | Martín-Blázquez A, Jiménez-Luna C, Díaz C, Martínez-Galán J, Prados J, Vicente F, Melguizo C, Genilloud O, Pérez Del Palacio J, Caba O. Discovery of Pancreatic Adenocarcinoma Biomarkers by Untargeted Metabolomics. Cancers (Basel). 2020;12. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 14] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 38. | Luo X, Liu J, Wang H, Lu H. Metabolomics identified new biomarkers for the precise diagnosis of pancreatic cancer and associated tissue metastasis. Pharmacol Res. 2020;156:104805. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 36] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 39. | Moore HB, Culp-Hill R, Reisz JA, Lawson PJ, Sauaia A, Schulick RD, Del Chiaro M, Nydam TL, Moore EE, Hansen KC, D'Alessandro A. The metabolic time line of pancreatic cancer: Opportunities to improve early detection of adenocarcinoma. Am J Surg. 2019;218:1206-1212. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 15] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 40. | Gaiser RA, Pessia A, Ateeb Z, Davanian H, Fernández Moro C, Alkharaan H, Healy K, Ghazi S, Arnelo U, Valente R, Velagapudi V, Sällberg Chen M, Del Chiaro M. Integrated targeted metabolomic and lipidomic analysis: A novel approach to classifying early cystic precursors to invasive pancreatic cancer. Sci Rep. 2019;9:10208. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 15] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 41. | Zhang X, Shi X, Lu X, Li Y, Zhan C, Akhtar ML, Yang L, Bai Y, Zhao J, Wang Y, Yao Y, Nie H. Novel Metabolomics Serum Biomarkers for Pancreatic Ductal Adenocarcinoma by the Comparison of Pre-, Postoperative and Normal Samples. J Cancer. 2020;11:4641-4651. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 5] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 42. | Mayerle J, Kalthoff H, Reszka R, Kamlage B, Peter E, Schniewind B, González Maldonado S, Pilarsky C, Heidecke CD, Schatz P, Distler M, Scheiber JA, Mahajan UM, Weiss FU, Grützmann R, Lerch MM. Metabolic biomarker signature to differentiate pancreatic ductal adenocarcinoma from chronic pancreatitis. Gut. 2018;67:128-137. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 157] [Cited by in F6Publishing: 169] [Article Influence: 28.2] [Reference Citation Analysis (0)] |
| 43. | Macias RIR, Muñoz-Bellvís L, Sánchez-Martín A, Arretxe E, Martínez-Arranz I, Lapitz A, Gutiérrez ML, La Casta A, Alonso C, González LM, Avila MA, Martinez-Chantar ML, Castro RE, Bujanda L, Banales JM, Marin JJG. A Novel Serum Metabolomic Profile for the Differential Diagnosis of Distal Cholangiocarcinoma and Pancreatic Ductal Adenocarcinoma. Cancers (Basel). 2020;12. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 15] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 44. | Rho SY, Lee SG, Park M, Lee J, Lee SH, Hwang HK, Lee MJ, Paik YK, Lee WJ, Kang CM. Developing a preoperative serum metabolome-based recurrence-predicting nomogram for patients with resected pancreatic ductal adenocarcinoma. Sci Rep. 2019;9:18634. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 45. | Fest J, Vijfhuizen LS, Goeman JJ, Veth O, Joensuu A, Perola M, Männistö S, Ness-Jensen E, Hveem K, Haller T, Tonisson N, Mikkel K, Metspalu A, van Duijn CM, Ikram A, Stricker BH, Ruiter R, van Eijck CHJ, van Ommen GB, ʼt Hoen PAC. Search for Early Pancreatic Cancer Blood Biomarkers in Five European Prospective Population Biobanks Using Metabolomics. Endocrinology. 2019;160:1731-1742. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 16] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 46. | Plaks V, Koopman CD, Werb Z. Cancer. Circulating tumor cells. Science. 2013;341:1186-1188. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 444] [Cited by in F6Publishing: 464] [Article Influence: 42.2] [Reference Citation Analysis (0)] |
| 47. | Jin KT, Chen XY, Lan HR, Wang SB, Ying XJ, Abdi SM, Wang W, Hu ZM, Mou XZ. Current progress in the clinical use of circulating tumor cells as prognostic biomarkers. Cancer Cytopathol. 2019;127:739-749. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 15] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 48. | Gall TMH, Belete S, Khanderia E, Frampton AE, Jiao LR. Circulating Tumor Cells and Cell-Free DNA in Pancreatic Ductal Adenocarcinoma. Am J Pathol. 2019;189:71-81. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 44] [Cited by in F6Publishing: 41] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 49. | Hugenschmidt H, Labori KJ, Brunborg C, Verbeke CS, Seeberg LT, Schirmer CB, Renolen A, Borgen EF, Naume B, Wiedswang G. Circulating Tumor Cells are an Independent Predictor of Shorter Survival in Patients Undergoing Resection for Pancreatic and Periampullary Adenocarcinoma. Ann Surg. 2020;271:549-558. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 35] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 50. | Court CM, Ankeny JS, Sho S, Winograd P, Hou S, Song M, Wainberg ZA, Girgis MD, Graeber TG, Agopian VG, Tseng HR, Tomlinson JS. Circulating Tumor Cells Predict Occult Metastatic Disease and Prognosis in Pancreatic Cancer. Ann Surg Oncol. 2018;25:1000-1008. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 50] [Cited by in F6Publishing: 58] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 51. | Effenberger KE, Schroeder C, Hanssen A, Wolter S, Eulenburg C, Tachezy M, Gebauer F, Izbicki JR, Pantel K, Bockhorn M. Improved Risk Stratification by Circulating Tumor Cell Counts in Pancreatic Cancer. Clin Cancer Res. 2018;24:2844-2850. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 50] [Cited by in F6Publishing: 57] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 52. | Varillas JI, Zhang J, Chen K, Barnes II, Liu C, George TJ, Fan ZH. Microfluidic Isolation of Circulating Tumor Cells and Cancer Stem-Like Cells from Patients with Pancreatic Ductal Adenocarcinoma. Theranostics. 2019;9:1417-1425. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 42] [Cited by in F6Publishing: 48] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 53. | Wei T, Zhang X, Zhang Q, Yang J, Chen Q, Wang J, Li X, Chen J, Ma T, Li G, Gao S, Lou J, Que R, Wang Y, Dang X, Zheng L, Liang T, Bai X. Vimentin-positive circulating tumor cells as a biomarker for diagnosis and treatment monitoring in patients with pancreatic cancer. Cancer Lett. 2019;452:237-243. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 46] [Cited by in F6Publishing: 72] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 54. | DiPardo BJ, Winograd P, Court CM, Tomlinson JS. Pancreatic cancer circulating tumor cells: applications for personalized oncology. Expert Rev Mol Diagn. 2018;18:809-820. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 55. | Pös O, Biró O, Szemes T, Nagy B. Circulating cell-free nucleic acids: characteristics and applications. Eur J Hum Genet. 2018;26:937-945. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 107] [Cited by in F6Publishing: 135] [Article Influence: 22.5] [Reference Citation Analysis (0)] |
| 56. | Moss J, Magenheim J, Neiman D, Zemmour H, Loyfer N, Korach A, Samet Y, Maoz M, Druid H, Arner P, Fu KY, Kiss E, Spalding KL, Landesberg G, Zick A, Grinshpun A, Shapiro AMJ, Grompe M, Wittenberg AD, Glaser B, Shemer R, Kaplan T, Dor Y. Comprehensive human cell-type methylation atlas reveals origins of circulating cell-free DNA in health and disease. Nat Commun. 2018;9:5068. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 331] [Cited by in F6Publishing: 475] [Article Influence: 79.2] [Reference Citation Analysis (0)] |
| 57. | Jaworski JJ, Morgan RD, Sivakumar S. Circulating Cell-Free Tumour DNA for Early Detection of Pancreatic Cancer. Cancers (Basel). 2020;12. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 12] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 58. | Alborelli I, Generali D, Jermann P, Cappelletti MR, Ferrero G, Scaggiante B, Bortul M, Zanconati F, Nicolet S, Haegele J, Bubendorf L, Aceto N, Scaltriti M, Mucci G, Quagliata L, Novelli G. Cell-free DNA analysis in healthy individuals by next-generation sequencing: a proof of concept and technical validation study. Cell Death Dis. 2019;10:534. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 65] [Cited by in F6Publishing: 66] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 59. | Guo J, Ma K, Bao H, Ma X, Xu Y, Wu X, Shao YW, Jiang M, Huang J. Quantitative characterization of tumor cell-free DNA shortening. BMC Genomics. 2020;21:473. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 60. | Shi J, Zhang R, Li J. Size profile of cell-free DNA: A beacon guiding the practice and innovation of clinical testing. Theranostics. 2020;10:4737-4748. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 29] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 61. | Lapin M, Oltedal S, Tjensvoll K, Buhl T, Smaaland R, Garresori H, Javle M, Glenjen NI, Abelseth BK, Gilje B, Nordgård O. Fragment size and level of cell-free DNA provide prognostic information in patients with advanced pancreatic cancer. J Transl Med. 2018;16:300. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 57] [Cited by in F6Publishing: 66] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 62. | Hamam HJ, Palaniyar N. Post-Translational Modifications in NETosis and NETs-Mediated Diseases. Biomolecules. 2019;9. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 54] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 63. | Kondo S, Sasaki M, Hosoi H, Sakamoto Y, Morizane C, Ueno H, Okusaka T. Incidence and risk factors for venous thromboembolism in patients with pretreated advanced pancreatic carcinoma. Oncotarget. 2018;9:16883-16890. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 22] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 64. | Hisada Y, Grover SP, Maqsood A, Houston R, Ay C, Noubouossie DF, Cooley BC, Wallén H, Key NS, Thålin C, Farkas ÁZ, Farkas VJ, Tenekedjiev K, Kolev K, Mackman N. Neutrophils and neutrophil extracellular traps enhance venous thrombosis in mice bearing human pancreatic tumors. Haematologica. 2020;105:218-225. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 64] [Cited by in F6Publishing: 96] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 65. | Hisada Y, Mackman N. Update from the laboratory: mechanistic studies of pathways of cancer-associated venous thrombosis using mouse models. Hematology Am Soc Hematol Educ Program. 2019;2019:182-186. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 66. | MANDEL P, METAIS P. Nuclear Acids In Human Blood Plasma. C R Seances Soc Biol Fil. 1948;142:241-243. [PubMed] [Cited in This Article: ] |
| 67. | Kruger S, Heinemann V, Ross C, Diehl F, Nagel D, Ormanns S, Liebmann S, Prinz-Bravin I, Westphalen CB, Haas M, Jung A, Kirchner T, von Bergwelt-Baildon M, Boeck S, Holdenrieder S. Repeated mutKRAS ctDNA measurements represent a novel and promising tool for early response prediction and therapy monitoring in advanced pancreatic cancer. Ann Oncol. 2018;29:2348-2355. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 75] [Cited by in F6Publishing: 80] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 68. | Watanabe F, Suzuki K, Tamaki S, Abe I, Endo Y, Takayama Y, Ishikawa H, Kakizawa N, Saito M, Futsuhara K, Noda H, Konishi F, Rikiyama T. Longitudinal monitoring of KRAS-mutated circulating tumor DNA enables the prediction of prognosis and therapeutic responses in patients with pancreatic cancer. PLoS One. 2019;14:e0227366. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 50] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 69. | Gai W, Sun K. Epigenetic Biomarkers in Cell-Free DNA and Applications in Liquid Biopsy. Genes (Basel). 2019;10. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 80] [Cited by in F6Publishing: 82] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 70. | Traube FR, Carell T. The chemistries and consequences of DNA and RNA methylation and demethylation. RNA Biol. 2017;14:1099-1107. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 68] [Cited by in F6Publishing: 84] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 71. | Shinjo K, Hara K, Nagae G, Umeda T, Katsushima K, Suzuki M, Murofushi Y, Umezu Y, Takeuchi I, Takahashi S, Okuno Y, Matsuo K, Ito H, Tajima S, Aburatani H, Yamao K, Kondo Y. A novel sensitive detection method for DNA methylation in circulating free DNA of pancreatic cancer. PLoS One. 2020;15:e0233782. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 14] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 72. | Li S, Wang L, Zhao Q, Wang Z, Lu S, Kang Y, Jin G, Tian J. Genome-Wide Analysis of Cell-Free DNA Methylation Profiling for the Early Diagnosis of Pancreatic Cancer. Front Genet. 2020;11:596078. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 13] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 73. | Guler GD, Ning Y, Ku CJ, Phillips T, McCarthy E, Ellison CK, Bergamaschi A, Collin F, Lloyd P, Scott A, Antoine M, Wang W, Chau K, Ashworth A, Quake SR, Levy S. Detection of early stage pancreatic cancer using 5-hydroxymethylcytosine signatures in circulating cell free DNA. Nat Commun. 2020;11:5270. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 89] [Cited by in F6Publishing: 76] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 74. | Cao F, Wei A, Hu X, He Y, Zhang J, Xia L, Tu K, Yuan J, Guo Z, Liu H, Xie D, Li A. Integrated epigenetic biomarkers in circulating cell-free DNA as a robust classifier for pancreatic cancer. Clin Epigenetics. 2020;12:112. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 21] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 75. | Cohen JD, Li L, Wang Y, Thoburn C, Afsari B, Danilova L, Douville C, Javed AA, Wong F, Mattox A, Hruban RH, Wolfgang CL, Goggins MG, Dal Molin M, Wang TL, Roden R, Klein AP, Ptak J, Dobbyn L, Schaefer J, Silliman N, Popoli M, Vogelstein JT, Browne JD, Schoen RE, Brand RE, Tie J, Gibbs P, Wong HL, Mansfield AS, Jen J, Hanash SM, Falconi M, Allen PJ, Zhou S, Bettegowda C, Diaz LA Jr, Tomasetti C, Kinzler KW, Vogelstein B, Lennon AM, Papadopoulos N. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science. 2018;359:926-930. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1752] [Cited by in F6Publishing: 1544] [Article Influence: 257.3] [Reference Citation Analysis (0)] |
| 76. | Bronkhorst AJ, Ungerer V, Holdenrieder S. The emerging role of cell-free DNA as a molecular marker for cancer management. Biomol Detect Quantif. 2019;17:100087. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 318] [Cited by in F6Publishing: 282] [Article Influence: 56.4] [Reference Citation Analysis (0)] |
| 77. | Zhang L, Liang Y, Li S, Zeng F, Meng Y, Chen Z, Liu S, Tao Y, Yu F. The interplay of circulating tumor DNA and chromatin modification, therapeutic resistance, and metastasis. Mol Cancer. 2019;18:36. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 35] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 78. | Kwapisz D. The first liquid biopsy test approved. Is it a new era of mutation testing for non-small cell lung cancer? Ann Transl Med. 2017;5:46. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 152] [Cited by in F6Publishing: 176] [Article Influence: 25.1] [Reference Citation Analysis (0)] |
| 79. | Bartel DP. Metazoan MicroRNAs. Cell. 2018;173:20-51. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2329] [Cited by in F6Publishing: 2222] [Article Influence: 370.3] [Reference Citation Analysis (0)] |
| 80. | O'Brien J, Hayder H, Zayed Y, Peng C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front Endocrinol (Lausanne). 2018;9:402. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2111] [Cited by in F6Publishing: 2546] [Article Influence: 424.3] [Reference Citation Analysis (0)] |
| 81. | Cui C, Cui Q. The relationship of human tissue microRNAs with those from body fluids. Sci Rep. 2020;10:5644. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 56] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 82. | Keller A, Fehlmann T, Backes C, Kern F, Gislefoss R, Langseth H, Rounge TB, Ludwig N, Meese E. Competitive learning suggests circulating miRNA profiles for cancers decades prior to diagnosis. RNA Biol. 2020;17:1416-1426. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 83. | Conti I, Varano G, Simioni C, Laface I, Milani D, Rimondi E, Neri LM. miRNAs as Influencers of Cell-Cell Communication in Tumor Microenvironment. Cells. 2020;9. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39] [Cited by in F6Publishing: 48] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 84. | Wang J, Chen J, Chang P, LeBlanc A, Li D, Abbruzzesse JL, Frazier ML, Killary AM, Sen S. MicroRNAs in plasma of pancreatic ductal adenocarcinoma patients as novel blood-based biomarkers of disease. Cancer Prev Res (Phila). 2009;2:807-813. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 392] [Cited by in F6Publishing: 418] [Article Influence: 27.9] [Reference Citation Analysis (0)] |
| 85. | Zimmerman JW, Burkhart RA, Chu NJ, Fertig EJ, Jaffee EM. Abstract 5716: miR-21 inhibition regulates mutant KRAS effector pathways and intercepts development of pancreatic ductal adenocarcinoma. Proceedings of the Annual Meeting of the American Association for Cancer Research 2020; 2020 Apr 27-28 and Jun 22-24; Philadelphia (PA). Cancer Res. 2020;80 Suppl 16:Abstract nr 5716. [Cited in This Article: ] |
| 86. | Vila-Navarro E, Duran-Sanchon S, Vila-Casadesús M, Moreira L, Ginès À, Cuatrecasas M, Lozano JJ, Bujanda L, Castells A, Gironella M. Novel Circulating miRNA Signatures for Early Detection of Pancreatic Neoplasia. Clin Transl Gastroenterol. 2019;10:e00029. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 32] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 87. | Cao Z, Liu C, Xu J, You L, Wang C, Lou W, Sun B, Miao Y, Liu X, Wang X, Zhang T, Zhao Y. Plasma microRNA panels to diagnose pancreatic cancer: Results from a multicenter study. Oncotarget. 2016;7:41575-41583. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 40] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 88. | Xu J, Cao Z, Liu W, You L, Zhou L, Wang C, Lou W, Sun B, Miao Y, Liu X, Zhang T, Zhao Y. Plasma miRNAs Effectively Distinguish Patients With Pancreatic Cancer From Controls: A Multicenter Study. Ann Surg. 2016;263:1173-1179. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 54] [Cited by in F6Publishing: 64] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 89. | Mazza T, Gioffreda D, Fontana A, Biagini T, Carella M, Palumbo O, Maiello E, Bazzocchi F, Andriulli A, Tavano F. Clinical Significance of Circulating miR-1273g-3p and miR-122-5p in Pancreatic Cancer. Front Oncol. 2020;10:44. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 15] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 90. | Liu G, Shao C, Li A, Zhang X, Guo X, Li J. Diagnostic Value of Plasma miR-181b, miR-196a, and miR-210 Combination in Pancreatic Cancer. Gastroenterol Res Pract. 2020;2020:6073150. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in F6Publishing: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 91. | Gablo N, Trachtova K, Prochazka V, Hlavsa J, Grolich T, Kiss I, Srovnal J, Rehulkova A, Lovecek M, Skalicky P, Berindan-Neagoe I, Kala Z, Slaby O. Identification and Validation of Circulating Micrornas as Prognostic Biomarkers in Pancreatic Ductal Adenocarcinoma Patients Undergoing Surgical Resection. J Clin Med. 2020;9. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 92. | Shams R, Saberi S, Zali M, Sadeghi A, Ghafouri-Fard S, Aghdaei HA. Identification of potential microRNA panels for pancreatic cancer diagnosis using microarray datasets and bioinformatics methods. Sci Rep. 2020;10:7559. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 37] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 93. | Liu G, Ji L, Ke M, Ou Z, Tang N, Li Y. miR-125a-3p is responsible for chemosensitivity in PDAC by inhibiting epithelial-mesenchymal transition via Fyn. Biomed Pharmacother. 2018;106:523-531. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 21] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 94. | Kandimalla R, Shimura T, Mallik S, Sonohara F, Tsai S, Evans DB, Kim SC, Baba H, Kodera Y, Von Hoff D, Chen X, Goel A. Identification of Serum miRNA Signature and Establishment of a Nomogram for Risk Stratification in Patients With Pancreatic Ductal Adenocarcinoma. Ann Surg. 2020;. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 9] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 95. | Oto J, Navarro S, Larsen AC, Solmoirago MJ, Plana E, Hervás D, Fernández-Pardo Á, España F, Kristensen SR, Thorlacius-Ussing O, Medina P. MicroRNAs and Neutrophil Activation Markers Predict Venous Thrombosis in Pancreatic Ductal Adenocarcinoma and Distal Extrahepatic Cholangiocarcinoma. Int J Mol Sci. 2020;21. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 26] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 96. | Yan Q, Hu D, Li M, Chen Y, Wu X, Ye Q, Wang Z, He L, Zhu J. The Serum MicroRNA Signatures for Pancreatic Cancer Detection and Operability Evaluation. Front Bioeng Biotechnol. 2020;8:379. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 13] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 97. | Valihrach L, Androvic P, Kubista M. Circulating miRNA analysis for cancer diagnostics and therapy. Mol Aspects Med. 2020;72:100825. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 55] [Cited by in F6Publishing: 47] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 98. | Xavier CPR, Caires HR, Barbosa MAG, Bergantim R, Guimarães JE, Vasconcelos MH. The Role of Extracellular Vesicles in the Hallmarks of Cancer and Drug Resistance. Cells. 2020;9. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 60] [Cited by in F6Publishing: 87] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 99. | Jurj A, Pop-Bica C, Slaby O, Ştefan CD, Cho WC, Korban SS, Berindan-Neagoe I. Tiny Actors in the Big Cellular World: Extracellular Vesicles Playing Critical Roles in Cancer. Int J Mol Sci. 2020;21. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 11] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 100. | Shehzad A, Islam SU, Shahzad R, Khan S, Lee YS. Extracellular vesicles in cancer diagnostics and therapeutics. Pharmacol Ther. 2021;223:107806. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 35] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 101. | van Niel G, D'Angelo G, Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol. 2018;19:213-228. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3060] [Cited by in F6Publishing: 4319] [Article Influence: 719.8] [Reference Citation Analysis (0)] |
| 102. | Stopa KB, Kusiak AA, Szopa MD, Ferdek PE, Jakubowska MA. Pancreatic Cancer and Its Microenvironment-Recent Advances and Current Controversies. Int J Mol Sci. 2020;21. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 23] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 103. | Nannan L, Oudart JB, Monboisse JC, Ramont L, Brassart-Pasco S, Brassart B. Extracellular Vesicle-Dependent Cross-Talk in Cancer-Focus on Pancreatic Cancer. Front Oncol. 2020;10:1456. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 9] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 104. | Capello M, Vykoukal JV, Katayama H, Bantis LE, Wang H, Kundnani DL, Aguilar-Bonavides C, Aguilar M, Tripathi SC, Dhillon DS, Momin AA, Peters H, Katz MH, Alvarez H, Bernard V, Ferri-Borgogno S, Brand R, Adler DG, Firpo MA, Mulvihill SJ, Molldrem JJ, Feng Z, Taguchi A, Maitra A, Hanash SM. Exosomes harbor B cell targets in pancreatic adenocarcinoma and exert decoy function against complement-mediated cytotoxicity. Nat Commun. 2019;10:254. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 98] [Cited by in F6Publishing: 97] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 105. | Zheng J, Hernandez JM, Doussot A, Bojmar L, Zambirinis CP, Costa-Silva B, van Beek EJAH, Mark MT, Molina H, Askan G, Basturk O, Gonen M, Kingham TP, Allen PJ, D'Angelica MI, DeMatteo RP, Lyden D, Jarnagin WR. Extracellular matrix proteins and carcinoembryonic antigen-related cell adhesion molecules characterize pancreatic duct fluid exosomes in patients with pancreatic cancer. HPB (Oxford). 2018;20:597-604. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 42] [Cited by in F6Publishing: 43] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 106. | Mikamori M, Yamada D, Eguchi H, Hasegawa S, Kishimoto T, Tomimaru Y, Asaoka T, Noda T, Wada H, Kawamoto K, Gotoh K, Takeda Y, Tanemura M, Mori M, Doki Y. MicroRNA-155 Controls Exosome Synthesis and Promotes Gemcitabine Resistance in Pancreatic Ductal Adenocarcinoma. Sci Rep. 2017;7:42339. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 176] [Cited by in F6Publishing: 179] [Article Influence: 25.6] [Reference Citation Analysis (0)] |
| 107. | Lai X, Wang M, McElyea SD, Sherman S, House M, Korc M. A microRNA signature in circulating exosomes is superior to exosomal glypican-1 Levels for diagnosing pancreatic cancer. Cancer Lett. 2017;393:86-93. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 192] [Cited by in F6Publishing: 234] [Article Influence: 33.4] [Reference Citation Analysis (0)] |
| 108. | Takahasi K, Iinuma H, Wada K, Minezaki S, Kawamura S, Kainuma M, Ikeda Y, Shibuya M, Miura F, Sano K. Usefulness of exosome-encapsulated microRNA-451a as a minimally invasive biomarker for prediction of recurrence and prognosis in pancreatic ductal adenocarcinoma. J Hepatobiliary Pancreat Sci. 2018;25:155-161. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 70] [Cited by in F6Publishing: 97] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 109. | Kawamura S, Iinuma H, Wada K, Takahashi K, Minezaki S, Kainuma M, Shibuya M, Miura F, Sano K. Exosome-encapsulated microRNA-4525, microRNA-451a and microRNA-21 in portal vein blood is a high-sensitive liquid biomarker for the selection of high-risk pancreatic ductal adenocarcinoma patients. J Hepatobiliary Pancreat Sci. 2019;26:63-72. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 39] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 110. | Kumar SR, Kimchi ET, Manjunath Y, Gajagowni S, Stuckel AJ, Kaifi JT. RNA cargos in extracellular vesicles derived from blood serum in pancreas associated conditions. Sci Rep. 2020;10:2800. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 31] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 111. | Kitagawa T, Taniuchi K, Tsuboi M, Sakaguchi M, Kohsaki T, Okabayashi T, Saibara T. Circulating pancreatic cancer exosomal RNAs for detection of pancreatic cancer. Mol Oncol. 2019;13:212-227. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 47] [Cited by in F6Publishing: 62] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 112. | Li J, Li Z, Jiang P, Peng M, Zhang X, Chen K, Liu H, Bi H, Liu X, Li X. Circular RNA IARS (circ-IARS) secreted by pancreatic cancer cells and located within exosomes regulates endothelial monolayer permeability to promote tumor metastasis. J Exp Clin Cancer Res. 2018;37:177. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 179] [Cited by in F6Publishing: 254] [Article Influence: 42.3] [Reference Citation Analysis (0)] |
| 113. | Seimiya T, Otsuka M, Iwata T, Tanaka E, Sekiba K, Shibata C, Moriyama M, Nakagawa R, Maruyama R, Koike K. Aberrant expression of a novel circular RNA in pancreatic cancer. J Hum Genet. 2021;66:181-191. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 17] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 114. | Castillo J, Bernard V, San Lucas FA, Allenson K, Capello M, Kim DU, Gascoyne P, Mulu FC, Stephens BM, Huang J, Wang H, Momin AA, Jacamo RO, Katz M, Wolff R, Javle M, Varadhachary G, Wistuba II, Hanash S, Maitra A, Alvarez H. Surfaceome profiling enables isolation of cancer-specific exosomal cargo in liquid biopsies from pancreatic cancer patients. Ann Oncol. 2018;29:223-229. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 121] [Cited by in F6Publishing: 150] [Article Influence: 30.0] [Reference Citation Analysis (0)] |
| 115. | Goto T, Fujiya M, Konishi H, Sasajima J, Fujibayashi S, Hayashi A, Utsumi T, Sato H, Iwama T, Ijiri M, Sakatani A, Tanaka K, Nomura Y, Ueno N, Kashima S, Moriichi K, Mizukami Y, Kohgo Y, Okumura T. An elevated expression of serum exosomal microRNA-191, - 21, -451a of pancreatic neoplasm is considered to be efficient diagnostic marker. BMC Cancer. 2018;18:116. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 141] [Cited by in F6Publishing: 158] [Article Influence: 26.3] [Reference Citation Analysis (0)] |
| 116. | Nakamura S, Sadakari Y, Ohtsuka T, Okayama T, Nakashima Y, Gotoh Y, Saeki K, Mori Y, Nakata K, Miyasaka Y, Onishi H, Oda Y, Goggins M, Nakamura M. Pancreatic Juice Exosomal MicroRNAs as Biomarkers for Detection of Pancreatic Ductal Adenocarcinoma. Ann Surg Oncol. 2019;26:2104-2111. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 55] [Cited by in F6Publishing: 51] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 117. | Han C, Seebacher NA, Hornicek FJ, Kan Q, Duan Z. Regulation of microRNAs function by circular RNAs in human cancer. Oncotarget. 2017;8:64622-64637. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 41] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 118. | Limb C, Liu DSK, Veno MT, Rees E, Krell J, Bagwan IN, Giovannetti E, Pandha H, Strobel O, Rockall TA, Frampton AE. The Role of Circular RNAs in Pancreatic Ductal Adenocarcinoma and Biliary-Tract Cancers. Cancers (Basel). 2020;12. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 119. | Aheget H, Tristán-Manzano M, Mazini L, Cortijo-Gutierrez M, Galindo-Moreno P, Herrera C, Martin F, Marchal JA, Benabdellah K. Exosome: A New Player in Translational Nanomedicine. J Clin Med. 2020;9. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 43] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 120. | Palazzolo S, Memeo L, Hadla M, Duzagac F, Steffan A, Perin T, Canzonieri V, Tuccinardi T, Caligiuri I, Rizzolio F. Cancer Extracellular Vesicles: Next-Generation Diagnostic and Drug Delivery Nanotools. Cancers (Basel). 2020;12. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 15] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 121. | Daoud AZ, Mulholland EJ, Cole G, McCarthy HO. MicroRNAs in Pancreatic Cancer: biomarkers, prognostic, and therapeutic modulators. BMC Cancer. 2019;19:1130. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 70] [Cited by in F6Publishing: 115] [Article Influence: 23.0] [Reference Citation Analysis (0)] |
| 122. | Landskron G, De la Fuente M, Thuwajit P, Thuwajit C, Hermoso MA. Chronic inflammation and cytokines in the tumor microenvironment. J Immunol Res. 2014;2014:149185. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1033] [Cited by in F6Publishing: 1085] [Article Influence: 108.5] [Reference Citation Analysis (0)] |
| 123. | Dunlop RJ, Campbell CW. Cytokines and advanced cancer. J Pain Symptom Manage. 2000;20:214-232. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 104] [Cited by in F6Publishing: 91] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 124. | West NR. Coordination of Immune-Stroma Crosstalk by IL-6 Family Cytokines. Front Immunol. 2019;10:1093. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 56] [Cited by in F6Publishing: 60] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 125. | Padoan A, Plebani M, Basso D. Inflammation and Pancreatic Cancer: Focus on Metabolism, Cytokines, and Immunity. Int J Mol Sci. 2019;20. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 118] [Cited by in F6Publishing: 183] [Article Influence: 36.6] [Reference Citation Analysis (0)] |
| 126. | Greer JB, Whitcomb DC. Inflammation and pancreatic cancer: an evidence-based review. Curr Opin Pharmacol. 2009;9:411-418. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 114] [Cited by in F6Publishing: 117] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 127. | Nielsen MF, Mortensen MB, Detlefsen S. Key players in pancreatic cancer-stroma interaction: Cancer-associated fibroblasts, endothelial and inflammatory cells. World J Gastroenterol. 2016;22:2678-2700. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 158] [Cited by in F6Publishing: 149] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 128. | Öhlund D, Handly-Santana A, Biffi G, Elyada E, Almeida AS, Ponz-Sarvise M, Corbo V, Oni TE, Hearn SA, Lee EJ, Chio II, Hwang CI, Tiriac H, Baker LA, Engle DD, Feig C, Kultti A, Egeblad M, Fearon DT, Crawford JM, Clevers H, Park Y, Tuveson DA. Distinct populations of inflammatory fibroblasts and myofibroblasts in pancreatic cancer. J Exp Med. 2017;214:579-596. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1020] [Cited by in F6Publishing: 1380] [Article Influence: 197.1] [Reference Citation Analysis (0)] |
| 129. | Apte M, Pirola RC, Wilson JS. Pancreatic stellate cell: physiologic role, role in fibrosis and cancer. Curr Opin Gastroenterol. 2015;31:416-423. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 87] [Cited by in F6Publishing: 81] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 130. | Provenzano PP, Cuevas C, Chang AE, Goel VK, Von Hoff DD, Hingorani SR. Enzymatic targeting of the stroma ablates physical barriers to treatment of pancreatic ductal adenocarcinoma. Cancer Cell. 2012;21:418-429. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1408] [Cited by in F6Publishing: 1468] [Article Influence: 122.3] [Reference Citation Analysis (0)] |
| 131. | Ansari D, Carvajo M, Bauden M, Andersson R. Pancreatic cancer stroma: controversies and current insights. Scand J Gastroenterol. 2017;52:641-646. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 34] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 132. | Erkan M, Hausmann S, Michalski CW, Fingerle AA, Dobritz M, Kleeff J, Friess H. The role of stroma in pancreatic cancer: diagnostic and therapeutic implications. Nat Rev Gastroenterol Hepatol. 2012;9:454-467. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 426] [Cited by in F6Publishing: 454] [Article Influence: 37.8] [Reference Citation Analysis (0)] |
| 133. | Foucher ED, Ghigo C, Chouaib S, Galon J, Iovanna J, Olive D. Pancreatic Ductal Adenocarcinoma: A Strong Imbalance of Good and Bad Immunological Cops in the Tumor Microenvironment. Front Immunol. 2018;9:1044. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 66] [Cited by in F6Publishing: 91] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 134. | Guerra C, Collado M, Navas C, Schuhmacher AJ, Hernández-Porras I, Cañamero M, Rodriguez-Justo M, Serrano M, Barbacid M. Pancreatitis-induced inflammation contributes to pancreatic cancer by inhibiting oncogene-induced senescence. Cancer Cell. 2011;19:728-739. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 363] [Cited by in F6Publishing: 369] [Article Influence: 28.4] [Reference Citation Analysis (0)] |
| 135. | Mace TA, Ameen Z, Collins A, Wojcik S, Mair M, Young GS, Fuchs JR, Eubank TD, Frankel WL, Bekaii-Saab T, Bloomston M, Lesinski GB. Pancreatic cancer-associated stellate cells promote differentiation of myeloid-derived suppressor cells in a STAT3-dependent manner. Cancer Res. 2013;73:3007-3018. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 264] [Cited by in F6Publishing: 341] [Article Influence: 31.0] [Reference Citation Analysis (0)] |
| 136. | Friess H, Berberat P, Schilling M, Kunz J, Korc M, Büchler MW. Pancreatic cancer: the potential clinical relevance of alterations in growth factors and their receptors. J Mol Med (Berl). 1996;74:35-42. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 93] [Cited by in F6Publishing: 98] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 137. | Cook N, Frese KK, Moore M. Assessing the role of the EGF receptor in the development and progression of pancreatic cancer. Gastrointest Cancer. 2014;4:23-37. [DOI] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in F6Publishing: 3] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 138. | Oliveira-Cunha M, Newman WG, Siriwardena AK. Epidermal growth factor receptor in pancreatic cancer. Cancers (Basel). 2011;3:1513-1526. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 86] [Cited by in F6Publishing: 103] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 139. | McCaffery I, Tudor Y, Deng H, Tang R, Suzuki S, Badola S, Kindler HL, Fuchs CS, Loh E, Patterson SD, Chen L, Gansert JL. Putative predictive biomarkers of survival in patients with metastatic pancreatic adenocarcinoma treated with gemcitabine and ganitumab, an IGF1R inhibitor. Clin Cancer Res. 2013;19:4282-4289. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 49] [Cited by in F6Publishing: 51] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 140. | Dahlem C, Barghash A, Puchas P, Haybaeck J, Kessler SM. The Insulin-Like Growth Factor 2 mRNA Binding Protein IMP2/IGF2BP2 is Overexpressed and Correlates with Poor Survival in Pancreatic Cancer. Int J Mol Sci. 2019;20. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 50] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 141. | Ahmed S, Bradshaw AD, Gera S, Dewan MZ, Xu R. The TGF-β/Smad4 Signaling Pathway in Pancreatic Carcinogenesis and Its Clinical Significance. J Clin Med. 2017;6. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 89] [Cited by in F6Publishing: 113] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 142. | Sabbadini F, Bertolini M, Matteis S, De Mangiameli D, Contarelli S, Pietrobono S, Melisi D. The Multifaceted Role of TGF- β in Gastrointestinal Tumors. Cancers (Basel). 2021;13:3960. [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 16] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 143. | Sakorafas GH, Tsiotou AG, Tsiotos GG. Molecular biology of pancreatic cancer; oncogenes, tumour suppressor genes, growth factors, and their receptors from a clinical perspective. Cancer Treat Rev. 2000;26:29-52. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 57] [Cited by in F6Publishing: 59] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 144. | Kalluri R, Zeisberg M. Fibroblasts in cancer. Nat Rev Cancer. 2006;6:392-401. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3347] [Cited by in F6Publishing: 3370] [Article Influence: 187.2] [Reference Citation Analysis (0)] |
| 145. | Kang X, Lin Z, Xu M, Pan J, Wang ZW. Deciphering role of FGFR signalling pathway in pancreatic cancer. Cell Prolif. 2019;52:e12605. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 32] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 146. | Rizwani W, Allen AE, Trevino JG. Hepatocyte Growth Factor from a Clinical Perspective: A Pancreatic Cancer Challenge. Cancers (Basel). 2015;7:1785-1805. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 18] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 147. | Kirby RE, Lewandrowski KB, Southern JF, Compton CC, Warshaw AL. Relation of epidermal growth factor receptor and estrogen receptor-independent pS2 protein to the malignant transformation of mucinous cystic neoplasms of the pancreas. Arch Surg. 1995;130:69-72. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 17] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 148. | Roshani R, McCarthy F, Hagemann T. Inflammatory cytokines in human pancreatic cancer. Cancer Lett. 2014;345:157-163. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 90] [Cited by in F6Publishing: 92] [Article Influence: 8.4] [Reference Citation Analysis (1)] |
| 149. | Yuan C, Rubinson DA, Qian ZR, Wu C, Kraft P, Bao Y, Ogino S, Ng K, Clancy TE, Swanson RS, Gorman MJ, Brais LK, Li T, Stampfer MJ, Hu FB, Giovannucci EL, Kulke MH, Fuchs CS, Wolpin BM. Survival among patients with pancreatic cancer and long-standing or recent-onset diabetes mellitus. J Clin Oncol. 2015;33:29-35. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 60] [Cited by in F6Publishing: 71] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 150. | Kim HW, Lee JC, Paik KH, Kang J, Kim J, Hwang JH. Serum interleukin-6 is associated with pancreatic ductal adenocarcinoma progression pattern. Medicine (Baltimore). 2017;96:e5926. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 32] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 151. | Ramsey ML, Talbert E, Ahn D, Bekaii-Saab T, Badi N, Bloomston PM, Conwell DL, Cruz-Monserrate Z, Dillhoff M, Farren MR, Hinton A, Krishna SG, Lesinski GB, Mace T, Manilchuk A, Noonan A, Pawlik TM, Rajasekera PV, Schmidt C, Guttridge D, Hart PA. Circulating interleukin-6 is associated with disease progression, but not cachexia in pancreatic cancer. Pancreatology. 2019;19:80-87. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 19] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 152. | Roussos ET, Condeelis JS, Patsialou A. Chemotaxis in cancer. Nat Rev Cancer. 2011;11:573-587. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 673] [Cited by in F6Publishing: 626] [Article Influence: 48.2] [Reference Citation Analysis (0)] |
| 153. | Torres C, Linares A, Alejandre MJ, Palomino-Morales RJ, Caba O, Prados J, Aránega A, Delgado JR, Irigoyen A, Martínez-Galán J, Ortuño FM, Rojas I, Perales S. Prognosis Relevance of Serum Cytokines in Pancreatic Cancer. Biomed Res Int. 2015;2015:518284. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 13] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 154. | Torres C, Perales S, Alejandre MJ, Iglesias J, Palomino RJ, Martin M, Caba O, Prados JC, Aránega A, Delgado JR, Irigoyen A, Ortuño FM, Rojas I, Linares A. Serum cytokine profile in patients with pancreatic cancer. Pancreas. 2014;43:1042-1049. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 33] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 155. | Le Naour F, Misek DE, Krause MC, Deneux L, Giordano TJ, Scholl S, Hanash SM. Proteomics-based identification of RS/DJ-1 as a novel circulating tumor antigen in breast cancer. Clin Cancer Res. 2001;7:3328-3335. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2] [Cited by in F6Publishing: 2] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 156. | Hu R, Lu C, Mostaghel EA, Yegnasubramanian S, Gurel M, Tannahill C, Edwards J, Isaacs WB, Nelson PS, Bluemn E, Plymate SR, Luo J. Distinct transcriptional programs mediated by the ligand-dependent full-length androgen receptor and its splice variants in castration-resistant prostate cancer. Cancer Res. 2012;72:3457-3462. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 418] [Cited by in F6Publishing: 448] [Article Influence: 37.3] [Reference Citation Analysis (0)] |
| 157. | Maacke H, Hundertmark C, Miska S, Voss M, Kalthoff H, Stürzbecher HW. Autoantibodies in sera of pancreatic cancer patients identify recombination factor Rad51 as a tumour-associated antigen. J Cancer Res Clin Oncol. 2002;128:219-222. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 33] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 158. | Dumstrei K, Chen H, Brenner H. A systematic review of serum autoantibodies as biomarkers for pancreatic cancer detection. Oncotarget. 2016;7:11151-11164. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 30] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 159. | Chen H, Werner S, Tao S, Zörnig I, Brenner H. Blood autoantibodies against tumor-associated antigens as biomarkers in early detection of colorectal cancer. Cancer Lett. 2014;346:178-187. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 63] [Cited by in F6Publishing: 65] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 160. | Zaenker P, Gray ES, Ziman MR. Autoantibody Production in Cancer--The Humoral Immune Response toward Autologous Antigens in Cancer Patients. Autoimmun Rev. 2016;15:477-483. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 133] [Cited by in F6Publishing: 141] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 161. | Gnjatic S, Ritter E, Büchler MW, Giese NA, Brors B, Frei C, Murray A, Halama N, Zörnig I, Chen YT, Andrews C, Ritter G, Old LJ, Odunsi K, Jäger D. Seromic profiling of ovarian and pancreatic cancer. Proc Natl Acad Sci U S A. 2010;107:5088-5093. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 143] [Cited by in F6Publishing: 150] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 162. | Capello M, Cappello P, Linty FC, Chiarle R, Sperduti I, Novarino A, Salacone P, Mandili G, Naccarati A, Sacerdote C, Beghelli S, Bersani S, Barbi S, Bassi C, Scarpa A, Nisticò P, Giovarelli M, Vineis P, Milella M, Novelli F. Autoantibodies to Ezrin are an early sign of pancreatic cancer in humans and in genetically engineered mouse models. J Hematol Oncol. 2013;6:67. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 38] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 163. | Tomaino B, Cappello P, Capello M, Fredolini C, Sperduti I, Migliorini P, Salacone P, Novarino A, Giacobino A, Ciuffreda L, Alessio M, Nisticò P, Scarpa A, Pederzoli P, Zhou W, Petricoin Iii EF, Liotta LA, Giovarelli M, Milella M, Novelli F. Circulating autoantibodies to phosphorylated α-enolase are a hallmark of pancreatic cancer. J Proteome Res. 2011;10:105-112. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 86] [Cited by in F6Publishing: 93] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 164. | Ghassem-Zadeh S, Hufnagel K, Bauer A, Frossard JL, Yoshida M, Kutsumi H, Acha-Orbea H, Neulinger-Muñoz M, Vey J, Eckert C, Strobel O, Hoheisel JD, Felix K. Novel Autoantibody Signatures in Sera of Patients with Pancreatic Cancer, Chronic Pancreatitis and Autoimmune Pancreatitis: A Protein Microarray Profiling Approach. Int J Mol Sci. 2020;21. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 4] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 165. | Hamanaka Y, Suehiro Y, Fukui M, Shikichi K, Imai K, Hinoda Y. Circulating anti-MUC1 IgG antibodies as a favorable prognostic factor for pancreatic cancer. Int J Cancer. 2003;103:97-100. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 130] [Cited by in F6Publishing: 138] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 166. | Tomaino B, Cappello P, Capello M, Fredolini C, Ponzetto A, Novarino A, Ciuffreda L, Bertetto O, De Angelis C, Gaia E, Salacone P, Milella M, Nisticò P, Alessio M, Chiarle R, Giuffrida MG, Giovarelli M, Novelli F. Autoantibody signature in human ductal pancreatic adenocarcinoma. J Proteome Res. 2007;6:4025-4031. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 58] [Cited by in F6Publishing: 60] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 167. | Lefrançais E, Ortiz-Muñoz G, Caudrillier A, Mallavia B, Liu F, Sayah DM, Thornton EE, Headley MB, David T, Coughlin SR, Krummel MF, Leavitt AD, Passegué E, Looney MR. The lung is a site of platelet biogenesis and a reservoir for haematopoietic progenitors. Nature. 2017;544:105-109. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 752] [Cited by in F6Publishing: 684] [Article Influence: 97.7] [Reference Citation Analysis (0)] |
| 168. | Wurdinger T, In 't Veld SGJG, Best MG. Platelet RNA as Pan-Tumor Biomarker for Cancer Detection. Cancer Res. 2020;80:1371-1373. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 25] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 169. | Best MG, Sol N, Kooi I, Tannous J, Westerman BA, Rustenburg F, Schellen P, Verschueren H, Post E, Koster J, Ylstra B, Ameziane N, Dorsman J, Smit EF, Verheul HM, Noske DP, Reijneveld JC, Nilsson RJA, Tannous BA, Wesseling P, Wurdinger T. RNA-Seq of Tumor-Educated Platelets Enables Blood-Based Pan-Cancer, Multiclass, and Molecular Pathway Cancer Diagnostics. Cancer Cell. 2015;28:666-676. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 569] [Cited by in F6Publishing: 548] [Article Influence: 60.9] [Reference Citation Analysis (0)] |
| 170. | Sol N, Wurdinger T. Platelet RNA signatures for the detection of cancer. Cancer Metastasis Rev. 2017;36:263-272. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 54] [Cited by in F6Publishing: 59] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 171. | Best MG, Wesseling P, Wurdinger T. Tumor-Educated Platelets as a Noninvasive Biomarker Source for Cancer Detection and Progression Monitoring. Cancer Res. 2018;78:3407-3412. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 79] [Cited by in F6Publishing: 140] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 172. | Braun A, Anders HJ, Gudermann T, Mammadova-Bach E. Platelet-Cancer Interplay: Molecular Mechanisms and New Therapeutic Avenues. Front Oncol. 2021;11:665534. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 42] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 173. | Campello E, Ilich A, Simioni P, Key NS. The relationship between pancreatic cancer and hypercoagulability: a comprehensive review on epidemiological and biological issues. Br J Cancer. 2019;121:359-371. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 43] [Cited by in F6Publishing: 58] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 174. | Khorana AA, Ahrendt SA, Ryan CK, Francis CW, Hruban RH, Hu YC, Hostetter G, Harvey J, Taubman MB. Tissue factor expression, angiogenesis, and thrombosis in pancreatic cancer. Clin Cancer Res. 2007;13:2870-2875. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 260] [Cited by in F6Publishing: 270] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 175. | AmraneDjedidi R, Rousseau A, Larsen AK, Elalamy I, Van Dreden P, Gerotziafas GT. Extracellular vesicles derived from pancreatic cancer cells BXPC3 or breast cancer cells MCF7 induce a permanent procoagulant shift to endothelial cells. Thromb Res. 2020;187:170-179. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 4] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 176. | Nadir Y, Brenner B. Heparanase procoagulant activity in cancer progression. Thromb Res. 2016;140 Suppl 1:S44-S48. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 27] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 177. | Ay C, Pabinger I, Cohen AT. Cancer-associated venous thromboembolism: Burden, mechanisms, and management. Thromb Haemost. 2017;117:219-230. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 208] [Cited by in F6Publishing: 264] [Article Influence: 33.0] [Reference Citation Analysis (0)] |
| 178. | Chen Y, Wang YR, Deng GC, Dai GH. CA19-9 decrease and survival according to platelet level in patients with advanced pancreatic cancer. BMC Cancer. 2019;19:860. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 8] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 179. | Xu ZS, Zhang FP, Zhang Y, Ou-Yang YP, Yu XW, Wang WL, Xu WJ, Luo ZQ. Prognostic role of the pre-treatment platelet-lymphocyte ratio in pancreatic cancer: a meta-analysis. Oncotarget. 2017;8:99003-99012. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 14] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 180. | Li W, Chen Y, Wang X, Shi Y, Dai G, Li X. Pretreatment platelet to lymphocyte ratio is predictive of overall survival in metastatic pancreatic ductal adenocarcinoma. Transl Cancer Res. 2019;8:17-22. [DOI] [Cited in This Article: ] [Cited by in Crossref: 1] [Cited by in F6Publishing: 1] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 181. | Shen XM, Xia YY, Lian L, Zhou C, Li XL, Han SG, Zheng Y, Gong FR, Tao M, Mao ZQ, Li W. Mean platelet volume provides beneficial diagnostic and prognostic information for patients with resectable gastric cancer. Oncol Lett. 2016;12:2501-2506. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 39] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 182. | Kumagai S, Tokuno J, Ueda Y, Marumo S, Shoji T, Nishimura T, Fukui M, Huang CL. Prognostic significance of preoperative mean platelet volume in resected non-small-cell lung cancer. Mol Clin Oncol. 2015;3:197-201. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 64] [Cited by in F6Publishing: 73] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 183. | Yin JB, Wang X, Zhang X, Liu L, Wang RT. Mean platelet volume predicts survival in pancreatic cancer patients with synchronous liver metastases. Sci Rep. 2018;8:6014. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 18] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 184. | Wang S, Li Z, Xu R. Human Cancer and Platelet Interaction, a Potential Therapeutic Target. Int J Mol Sci. 2018;19. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 43] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 185. | Yan M, Jurasz P. The role of platelets in the tumor microenvironment: From solid tumors to leukemia. Biochim Biophys Acta. 2016;1863:392-400. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 97] [Cited by in F6Publishing: 108] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 186. | Mantini G, Meijer LL, Glogovitis I, In 't Veld SGJG, Paleckyte R, Capula M, Le Large TYS, Morelli L, Pham TV, Piersma SR, Frampton AE, Jimenez CR, Kazemier G, Koppers-Lalic D, Wurdinger T, Giovannetti E. Omics Analysis of Educated Platelets in Cancer and Benign Disease of the Pancreas. Cancers (Basel). 2020;13. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 14] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 187. | Nassa G, Giurato G, Cimmino G, Rizzo F, Ravo M, Salvati A, Nyman TA, Zhu Y, Vesterlund M, Lehtiö J, Golino P, Weisz A, Tarallo R. Splicing of platelet resident pre-mRNAs upon activation by physiological stimuli results in functionally relevant proteome modifications. Sci Rep. 2018;8:498. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 46] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 188. | Sunderland N, Skroblin P, Barwari T, Huntley RP, Lu R, Joshi A, Lovering RC, Mayr M. MicroRNA Biomarkers and Platelet Reactivity: The Clot Thickens. Circ Res. 2017;120:418-435. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 126] [Cited by in F6Publishing: 132] [Article Influence: 18.9] [Reference Citation Analysis (0)] |
| 189. | Koppers-Lalic D, Hackenberg M, de Menezes R, Misovic B, Wachalska M, Geldof A, Zini N, de Reijke T, Wurdinger T, Vis A, van Moorselaar J, Pegtel M, Bijnsdorp I. Noninvasive prostate cancer detection by measuring miRNA variants (isomiRs) in urine extracellular vesicles. Oncotarget. 2016;7:22566-22578. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 93] [Cited by in F6Publishing: 99] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 190. | Zhang YH, Huang T, Chen L, Xu Y, Hu Y, Hu LD, Cai Y, Kong X. Identifying and analyzing different cancer subtypes using RNA-seq data of blood platelets. Oncotarget. 2017;8:87494-87511. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 35] [Article Influence: 5.0] [Reference Citation Analysis (0)] |