Published online Jun 26, 2015. doi: 10.4252/wjsc.v7.i5.823
Peer-review started: December 6, 2014
First decision: February 7, 2015
Revised: March 5, 2015
Accepted: April 1, 2015
Article in press: April 7, 2015
Published online: June 26, 2015
Pathogenic mutations involving DNA repeat expansions are responsible for over 20 different neuronal and neuromuscular diseases. All result from expanded tracts of repetitive DNA sequences (mostly microsatellites) that become unstable beyond a critical length when transmitted across generations. Nearly all are inherited as autosomal dominant conditions and are typically associated with anticipation. Pathologic unstable repeat expansions can be classified according to their length, repeat sequence, gene location and underlying pathologic mechanisms. This review summarizes the current contribution of mutant pluripotent stem cells (diseased human embryonic stem cells and patient-derived induced pluripotent stem cells) to the research of unstable repeat pathologies by focusing on particularly large unstable noncoding expansions. Among this class of disorders are Fragile X syndrome and Fragile X-associated tremor/ataxia syndrome, myotonic dystrophy type 1 and myotonic dystrophy type 2, Friedreich ataxia and C9 related amyotrophic lateral sclerosis and/or frontotemporal dementia, Facioscapulohumeral Muscular Dystrophy and potentially more. Common features that are typical to this subclass of conditions are RNA toxic gain-of-function, epigenetic loss-of-function, toxic repeat-associated non-ATG translation and somatic instability. For each mechanism we summarize the currently available stem cell based models, highlight how they contributed to better understanding of the related mechanism, and discuss how they may be utilized in future investigations.
Core tip: This review summarizes the current contribution of mutant pluripotent stem cells (diseased HESCs and patient-derived induced pluripotent stem cells) to the research of unstable repeat pathologies by focusing on particularly large unstable noncoding expansions. It demonstrates their importance as an unlimited cell source for generating rarely available impaired cells in culture, and as a model system for exploring the mechanisms that are involved with this class of mutations. For each mechanism we describe the currently available stem cell based models, highlight how they contributed to better understanding of the related mechanism, and discuss how they may be utilized in future investigations.
- Citation: Yanovsky-Dagan S, Mor-Shaked H, Eiges R. Modeling diseases of noncoding unstable repeat expansions using mutant pluripotent stem cells. World J Stem Cells 2015; 7(5): 823-838
- URL: https://www.wjgnet.com/1948-0210/full/v7/i5/823.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v7.i5.823
Unstable repeat expansions, termed also dynamic mutations, are the cause of over 20 different neurodevelopmental, neurodegenerative and neuromuscular heritable conditions that result from a change in the number of repeat units in coding genes when ordered in tandem (for comprehensive review see[1]). Repeat instabilities change across generations, and may vary between cells within affected individuals. Nearly all are inherited as autosomal dominant conditions and are typically associated with anticipation, meaning that the disease symptoms tend to be more severe and occur earlier in age with successive generations. Most unstable repeat associated disorders are caused by the addition of repeat units (mostly tri-nucleotides) that, above a certain threshold, lead to misregulation of genes and/or to their RNA/protein products. Disease associated unstable repeat expansions can be classified into two groups according to the length and gene location of the expansion. One class of disorders results from small repeat copy changes (few to tens of units) that usually reside in exon coding regions of genes. Among them are the ployglutamine CAG expansion disorders like Huntington disease, Huntington’s disease like 2 (HDL2), Dentatorubral-pallidoluysian atrophy (DRPLA), Spinal and bulbar muscular atrophy (SBMA) and spinocerebellar ataxia (SCA) 1, 2, 3, 6, 7 and 17, which lead to protein gain-of-function alterations. A second class of unstable repeat expansion disorders results from large expansions, ranging from hundreds to thousands of copies. Large pathogenic repeat expansions are typically located in noncoding regions including promoters, introns and untranslated regions (UTRs) of genes[2]. Among disorders in this class are Fragile X syndrome (FXS) and [Fragile X-associated tremor/ataxia syndrome (FXTAS); both FXS and FXTAS are caused by large expansion of a CGG repeat in the 5’-UTR of the FMR1 gene[3-5]], [myotonic dystrophy type 1 (DM1); caused by a CTG expansion in the 3’UTR of the DMPK gene[6-8]] and type 2 (DM2; caused by a CCTG expansion in intron 1 of the ZNF9 gene[9]), [Friedreich ataxia (FRDA); caused by a GAA expansion in intron 1 of the Frataxin gene[10]], C9 related [amyotrophic lateral sclerosis and/or frontotemporal dementia (ALS-FTD); caused by a GGGGCC[11] expansion in intron 1 of the C9orf72 gene[12]], and [Facioscapulohumeral Muscular Dystrophy (FSHD); caused by a contraction of the D4Z4 macrosatellite repeat in sub-telomeres of chromosome 4q35].
The outcome of the expansion mutation may be different depending on its gene location and length. Unlike small expansions, which commonly result in alterations in protein function, large noncoding expansions introduce further complexity because they can lead to either loss-of-function, RNA gain-of-function, toxic protein gain-of-function, or to a combination of all these pathogenic mechanisms in unison[13]. In addition and in contrast to small expansions, large noncoding expansions frequently coincide with marked changes in repeat tract length between and within tissues of affected individuals[8,14,15]. This phenomenon, termed somatic repeat instability, results in mosaicism for expansion size and occasionally correlates with disease age of onset and severity. Modeling dynamic mutations, specifically large expansions, in mice can be particularly challenging due to the difficulty in artificially inducing and stably maintaining very large repeat expansions (especially CG-rich) in vitro and in vivo. In addition, despite the similarities between mouse and human, there are still major differences between the two species, leading to dissimilarities in biochemical pathways and phenotypes. These crucial discrepancies emphasize the need for complement model systems that will reproducibly copy the underlying mechanisms and clinical phenotypes in human.
Mutant human pluripotent stem cells, embryonic [human embryonic stem cells (HESCs)] or artificially induced [induced pluripotent stem cells (iPS cells)], may provide a great opportunity to study unstable repeat expansions by complementing existing models[16]. They are expected to be especially useful in the study of heritable conditions where the animal model fails to fully recapitulate the phenotype of the disease in human, or when disease relevant cell cultures from patients are unavailable. The many advantages to the utilization of these cells for biomedical research include the fact that they are human derived, untransformed cells with an unlimited self-renewing capacity. In addition, they have the potential to differentiate into a wide range of cell types in culture, and recapitulate early human embryo development while they differentiate in vitro. Moreover, as mutant pluripotent stem cells can be derived directly from genetically affected embryos that are obtained through preimplantation genetic diagnosis (PGD) procedures (HESC)[17], or by reprogramming somatic cells obtained from patients (iPS)[18], they can reproduce disease cellular phenotypes as they occur in vivo without the need to artificially intervene with their genome through genetic manipulation. In addition, they are human derived and potentially be used to generate large amounts of impaired disease relevant cells in culture. This is particularly beneficial in the case of unstable repeat pathologies, where studies are frequently limited to postmortem brain samples or to unsuitable cell types obtained from patients such as peripheral blood cells or skin fibroblasts. Furthermore, as these cells can recapitulate early stage embryo development, they may be particularly valuable in modeling disease associated mechanisms that are developmentally regulated such as those that are elicited by differentiation. In terms of applied research, mutant pluripotent stem cells provide a powerful cell culture based system for gene correction. For instance, they may facilitate the efficient induction of irreversible changes in DNA that will correct the disease causing mutation by shortening the repeat tract through genome editing or other gene manipulation approaches[19]. They can also provide a platform for drug screening and development, conditioned by the accessibility of efficient differentiation protocols and the availability of reliable biomarkers.
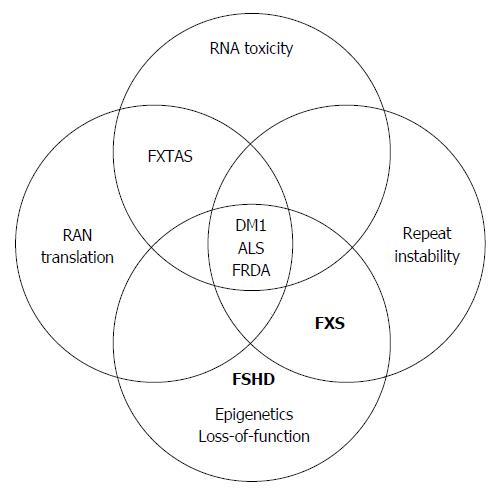
In this review we summarize the current contribution of mutant pluripotent stem cells to the research of unstable repeat pathologies by focusing on common mechanisms that are associated with large unstable noncoding expansions (Figure 1). A complete survey of the data regarding the use of mutant pluripotent stem cells for modeling phenotypes of coding unstable repeat expansions, and the use of these cells as a platform for gene therapy, are beyond the scope of this review and can be found elsewhere[20-22]. The discussion here will thus be limited to only pathological noncoding repeat expansions. For each mechanism we highlight the currently available pluripotent stem cell models by describing their exclusive utility for investigation of the mechanism, how they contributed to better understanding of the related mechanism, and we raise potential routes for future investigation.
Over the past decade it has become evident that RNAs containing abnormally large microsatellite repeat expansions underlie the pathogenesis of at least 9 dominantly inherited human diseases. These are mostly neurological and neuromuscular diseases, including DM1[23], DM2[24], FXTAS[25], C9/ALS-FTD[26], SCA3[27], SCA8[28], SCA10[29], SCA31[30] and HDL2[31]. These diseases arise when the repeat-containing RNA disrupts the function of specific RNA binding proteins (RBPs) in trans. The paradigm for such toxic RNA gain-of-function pathology is DM1, where a CUG repeat expansion located in the 3’-UTR of the DMPK gene interferes with the cellular function of specific CUG-binding splice-regulating proteins termed MBNL1 and CUG-BP1 (CLEF protein family)[32,33]. The altered function of these splice regulators hinders the normal processing of transcripts encoding over twenty different muscle genes, resulting in inappropriate expression of embryonic rather than adult splice variants in adult disease manifesting tissues[34,35]. The general hallmarks of unstable repeat disorders with RNA-mediated toxicity are the accumulation of repeat-containing RNAs in intra-nuclear inclusions along with the sequestration of common RBPs (mainly proteins involved with RNA metabolism) and alternative splicing misregulation[36]. Co-localization of the large ribonucleoprotein (RNP) aggregates with ubiquitin and proteasome subunits is a common feature[37]. These striking parallels have led to the identification of the most common (currently known) mechanisms underlying noncoding unstable microsatellite disorders. This subclass of disorders is caused not due to the loss of function of the mutant transcripts and haploinsufficiency, but rather from the loss of function of RBPs that aggregate with the lengthy RNAs, and possibly from the toxic effect of the aggregates themselves. As the RNA seeds accumulation of the proteins it interacts with, it alters their biological function by changing their abundance and/or localization in the cell. Furthermore, since most of these expansions are bi-directionally transcribed, multiple repeat-containing toxic RNAs seem to be involved[38-40]. The susceptibility of neurons to this process is a common feature, most likely related to the ageing course of this specific cell type. Considering the reversibility of the process[41-44], it may be possible to develop effective therapeutic strategies to ameliorate the symptoms of these diseases.
Taking advantage of two DM1 affected pluripotent stem cell lines derived from embryos identified through PGD, Denis et al[45] produced impaired early progenitor neural cells in culture. To validate the pathological relevance of their differentiated cells, they show that DM1-neural stem cells (NSCs) and derived neurons harbored intranuclear RNP foci, contain the mutant RNA and co-localize with the splice factor MBNL1. Interestingly, functional characterization of DM1 HESC-derived neurons revealed reduced proliferative capacity and increased autophagy linked to the mTOR signaling pathway. In addition, the researchers were able to demonstrate that the loss of function of MBNL1 in wild type (WT) HESCs results in alteration of the mTOR signaling pathway, whereas gain-of-function experiments rescued the phenotype. Collectively, their findings provided a mechanism by which the DM1 mutation might affect a major signaling pathway, and highlighted the pertinence of using pluripotent stem cells to study neuronal defects in general. A complement model system to support the formation of intranuclear RNA foci by in vitro derived neurons was developed in iPS cells, validating the potential of pluripotent stem cells to offer an unlimited cell resource for CNS mechanistic studies[46].
A different disease for which pluripotent stem cells have been employed to further understand the toxic RNA-mediated mechanism underlying unstable repeat expansions is in C9/ALS-FTD. Amyotrophic lateral sclerosis (ALS) and/or frontotemporal dementia (FTD) is an autosomal dominant neurodegenerative disorder characterized by adult onset of one or both of these conditions in an affected individual[47-49]. The leading cause for ALS-FTD is a G4C2 repeat expansion in the first intron of the C9orf72 gene, between noncoding exons 1a and 1b. Expansion of G4C2 repeat tract beyond a certain threshold, results in RNA gain-of-function resulting from bi-directional transcription[40,50]. Its low incidence (1:100000) and mainly sporadic occurrence (90%), late onset, and accelerated progression, have thus far facilitated the preparation of iPS cells, but not HESC lines. By establishing multiple iPS clones from skin biopsies of patients’ fibroblasts with over 1000 G4C2 repeats, researchers were able to generate motor neurons in culture that recapitulate key neuropathological features of ALS-FTD including RNA foci reminiscent of those observed in postmortem brain autopsies from patients[51]. Interestingly, the RNA foci containing G4C2 repeats were present in all iPSC-derived human neurons as well as in undifferentiated iPSCs and primary fibroblasts. This suggests that these foci are not restricted to a specific cell type or developmental stage, as one would predict if functionally significant. Moreover, transcription of the repeat containing C9orf72 transcripts, led to accumulation of G4C2 repeat-containing RNA foci selectively in various C9-ALS iPSC-derived neural cell types[52]. It was also shown that the pathologic RNA foci co-localize with the major known RBPs like hnRNPA1, Pur-α[52] and ADARB2[53], suggesting that they may be able to disrupt RNA metabolism. Furthermore, it was found that the in vitro derived C9-ALS motor neurons present altered expression of genes involved in membrane excitability, and demonstrated a diminished capacity to fire continuous spikes upon depolarization compared to a WT motor neuron control[52]. By targeting the RNA foci through antisense oligonucleotide (ASO), either through knockdown (KD) of the full length C9orf72 transcripts or by differentially targeting the repeat containing mRNA isoforms (V1 and V3), researchers were able to suppress RNA foci formation and reverse gene expression defects in C9-ALS neuronal cells[52,53], as well as, rescue to some extent disease phenotype (Glutamate toxicity[53]). These data show that patient-derived motor neurons can be used to elucidate the pathogenic role of the toxic RNA in C9/ALS-FTD. More generally, they neatly illustrate the power of mutant pluripotent cells as a translational platform for therapeutic development.
Many questions regarding this repeat-associated mechanism remain to be addressed. For example, what dictates the cell-type specificity of this process? How is it age related? What is the protein composition of the aggregates in each disorder and, what is the nature of the inclusions? Are they toxic or do they simply represent clearance of toxic RNA? In addition, although animal models have provided firm evidence for RNA gain-of-function as a major contributing mechanism[54,55], clinical and pathophysiological differences between human and available animals (mouse and Drosophila) limit their usefulness in delineating specific mechanisms underlying these toxic RNA-mediated pathologies[56-61]. Therefore, a complement human-based model system is needed to enable molecular characterization of the onset and course of this class of conditions.
Among the disease associated expansions there are several, counting FXS, FRDA, DM1, and C9/ALS-FTD, that results from particularly large noncoding expansions (hundreds to thousands of copies) that reside within CpG islands (CGIs). CGIs typically remains free of DNA methylation and are rich in histone modifications associated with transcriptionally active chromatin. Yet, expansion of repeat copy number beyond a certain threshold leads to local acquirement of abnormal CpG methylation in addition to loss of active histone modifications and concomitant gain of repressive histone modifications that are typical to densely packed chromatin (like H3K4 demethylation and H3K9 and H3K27 tri-methylation, respectively). These aberrant epigenetic modifications can change local gene transcription. Their effect may vary from one locus to another, depending on the location of the expansion in the gene. While in some genes it abolishes the activity of a promoter (as in FXS and C9orf72)[62,63] or an enhancer (as in DM1)[64], in other genes it interferes with RNA transcription elongation (as in FRDA)[65] or termination (as in FSHD[66]). Furthermore, even when hypermethylation leads to the loss of promoter activity, the outcome may be very different. For example, while in FXS transcriptional silencing results in FMRP protein deficiency (loss-of-protein function)[67], in C9/ALS-FTD epigenetic silencing may potentially lead to increased use of an alternative promoter (RNA gain-of-function)[39]. In addition, in some disorders hypermethylation seems to be acquired in a developmentally regulated process (FXS)[17] whereas in others it is more of a tissue or an age-dependent phenomenon (DM1)[68]. Therefore, it is essential to first understand when, where and how hypermethylation affects local gene expression, in order to examine whether and by what mechanism this contributes to disease pathogenesis. More generally, by unraveling how repeat expansions lead to the loss of CGI identity, we may gain new insights on how CpG islands are normally protected from de novo methylation throughout the genome.
The availability of mutant pluripotent stem cell lines carrying pathologic repeat expansions may provide a great model system to address the questions of when, where and how hypermethylation is aberrantly acquired. In particular, such cell lines can be utilized to better determine the timing, intervene with the mechanism, and monitor the effect of epigenetic modifications. In addition, they may provide an exceptional opportunity to rescue the mutant phenotype by resetting the undesired epigenetic modifications that are coupled with the expansion through cell reprogramming.
To date, pluripotent stem cell-based studies related to the epigenetic aspects of unstable repeat expansions have been mainly limited to FXS. In FXS, when the CGGs reach the full mutation range, they lead to the spread of abnormal CpG methylation and repressive histone modifications at the 5’ regulatory region, resulting in epigenetic gene silencing and consequently to FMRP protein deficiency in patients[62]. Initially, when the first FXS XY HESC line was established, it was found to express FMR1 at comparable levels with WT HESCs and to be completely FMR1 unmethylated[17]. These findings validated a commonly accepted view that FMR1 epigenetic silencing is a developmentally regulated process that is triggered by differentiation. Moreover, it encouraged investigators to propose that since epigenetic FMR1 inactivation is differentiation-dependent (rather than being maternally transmitted in the gamete), it may be possible to rescue the cellular phenotype of FXS neurons simply by removing the epigenetic marks that are erroneously gained, through cell de-differentiation in culture. However, as more FXS HESC lines were available, it became apparent that FMR1 hypermethylation commonly occurs in the undifferentiated state. Taking advantage of a large set of FXS HESCs (9 cell lines, including the former), Avitzour and colleagues found that FMR1 hypermethylation frequently occurs in these cell lines (six of nine lines, ranging from 24% to 65%)[69]. This suggests that the wide variability in methylation levels between and within the different FXS HESC lines reflects a widespread event within mutant FMR1 embryos, where methylation state is initially set. It should be interesting to determine whether FXS preimplantation embryos are already FMR1 methylated, and whether methylation at this stage is incomplete. Mechanistically, the researchers tried to address the question of how the CGG expansion at the FMR1 locus leads to de novo methylation of the CpG island at the 5’-UTR of FMR1[69]. They aimed to associate FMR1 hypermethylation with CTCF binding next to the repeats, taking in to consideration the well-known role of CTCF as an insulator binding protein that counteracts heterochromatin spreading[70,71] and taking in to account previous reports demonstrating CTCF occupancy next to the CGG repeats at the FMR1 locus in differentiated cells. However, since no enrichment for CTCF could be detected, in both WT and affected HESC lines, this study ruled out the possibility that hypermethylation may be attributed by the binding loss of CTCF, as formerly suggested[38,72]. Employing two other XY FXS HESC lines, Colak and colleagues uncovered an mRNA-mediated mechanism that drives epigenetic FMR1 inactivation in a way that relies on neuronal differentiation[73]. In this study the researchers showed that the CGG-lengthy FMR1 mRNA hybridizes to the complementary CGG-repeat portion of the FMR1 gene to form an RNA:DNA duplex. Upon differentiation the RNA:DNA hybrid elicits gene silencing on the protein and mRNA levels. According to their study, disrupting the interaction of the mRNA with the CGG-repeat portion of the FMR1 gene at a critical time point during cell differentiation prevents promoter silencing. It remains to be determined whether silencing in undifferentiated cells is achieved by the same mechanism or by distinct one, and whether the loss of FMR1 mRNA and protein in the FXS HESC-derived neurons involves DNA hypermethylation.
In line with this study, it has been proposed that it may be possible to reverse the adverse effect of the modifications and correct the mutant phenotype by reprogramming terminally differentiated FXS cells. However, somatic cell reprogramming of FXS patients’ fibroblasts by numerous independent studies failed to de-methylate and reactivate the gene simply by de-differentiation[74], suggesting that once established, methylation is irreversible and/or is acquired prior to blastocyst formation[75-77]. In line with this view, it should be feasible to obtain FXS-iPS clones that are FMR1 active from somatic cells with a full mutation provided that they carry an unmethylated full expansion at least in some of their cells. On the other hand, perhaps reprogramming may wrongly lead to hypermethylation of this mutation prone site, regardless of its initial epigenetic state. Indeed, reprogramming fibroblasts from an unusual asymptomatic subject, bearing a full mutation that is entirely unmethylated, resulted in complete silencing, regardless of the reprogramming method used[78]. Maybe the high levels of DNMT3b, that are induced as part of the reprogramming process[79], led to this unexpected result. This suggests that FXS iPS cells, and potentially additional iPS models for epigenetically regulated disorders, may not serve as an ideal model for better understanding the mechanism by which hypermethylation is induced. Reprogramming of somatic cells from additional subjects with hypomethylated expansions will be required in order to establish whether this is a widespread event that is irrelevant to the disease. Regardless, it should be possible to use these disease affected iPS cells to explore the consequence of FMRP deficiency or reverse this effect in in vitro differentiated neurons, a disease relevant cell type. Indeed, re-activation of FMR1 by the treatment with the de-methylating agent 5-azaC has already been successfully practiced in neuron derived FXS-iPS cells[80].
Apart from FXS HESCs, there is indirect evidence to suggest that hypermethylation is recapitulated in patient derived iPS cells for other unstable repeat related loci. One example is C9/ALS-FTD iPS cells that carry a G4C2 expansion at the C9orf72 gene. In the C9orf72 the G4C2 repetitive sequence is flanked by two CpG islands (CGIs) that span from the promoter sequence into intron 1 of C9orf72. Like many CGIs in the genome, this region typically remains unmethylated and is transcriptionally active. Yet, expansion to more than 90 repeats leads to the formation of one large CGI exclusively in patients, and to local acquirement of abnormal CpG methylation upstream[11]. Hypermethylation is coupled with the local gain of repressive histone modifications, and is responsible for the inactivation of variant 2 of C9orf72 that, unlike repeat-containing variants 1 and 3, initiates transcription from exon 1b and harbors the G4C2 sequence in its promoter[26]. Overall, C9/ALS-FTD iPS cells demonstrate altered gene expression. While some reports demonstrate general reduction in the transcription of all variants, others show differential expression favoring variants 1 and 3 over variant 2[26,52,81]. All together, this suggests that hypermethylation in fact exist in these cells. Nevertheless, the contribution of epigenetic gene silencing to disease pathogenesis is not fully clear. There is conflicting evidence regarding the correlation between hypermethylation and type of pathology (ALS vs FTD), age of onset or severity[11,82,83]. Experimental evidence in animal models demonstrates inconsistent results[84,85]. On the other hand, hypermethylation was suggested to have a protective role from the accumulation of pathogenic RNA/protein foci contributed by repeat-containing mRNA variants 1 and 3[86]. These conflicting studies warrant further investigation regarding the contribution of DNA methylation to ALS-FTD pathogenesis, and may be considerably advanced by the use of in vitro derived neurons from C9/ALS-FTD pluripotent cells.
Another elegant example for the power of pluripotent stem cells in resolving the contribution of epigenetics to disease pathogenesis is in FSHD. FSHD is caused by a contraction of a macrosatellite repeat (D4Z4 repeat, 3.3 kb unit) located in the sub-telomeres of chromosome 4q35. Non-affected individuals carry 11-100 repeat units, while FSHD patients have less than 10 repeats, but must have at least one unit to manifest the disease[87-89]. Interestingly, the loss of DNA methylation and reduced H3K9me3 that coincide with repeat contraction have been found to be causally related to the up-regulation of a full transcript from the DUX4 gene, a transcript which is exclusive to the most distal (telomeric) unit of the D4Z4 repeat containing region[66,80-93]. Expression of the full length DUX4 transcript, which is exclusive to FSHD myotubes, is responsible for the induction of nuclear foci, impaired myogenesis and ultimately to cell death by apoptosis[94]. Immediately downstream to the most-telomeric unit there is a polymorphic site that acts as an alternative polyadenylation signal that controls the length and stability of the DUX4 transcript. While normal alleles produce short transcripts and truncated proteins, contracted FSHD alleles utilize a cryptic splice donor to form a full length toxic protein. As alternative polyadenylation seemed to be a tissue-specific process, it was speculated to be a developmentally regulated event that is mediated by the loss of repressive epigenetic modifications due to D4Z4 repeat contraction. By comparing WT and FSHD iPS cell lines, Snider and colleagues[94] found that both cell types equally express full length DUX4, independent of repeat number. However, upon differentiation, full length expression was switched off and converted back to short transcripts in WT controls while full length expression persisted in the diseased cells. Importantly, the researchers were able to correlate the switch in splice site usage with the enhancement of repressive chromatin modifications (H3K9me3) in the WT differentiated cells. Thus, it appears that H3K9me3 modification (and most likely CpG methylation) mediates the splicing for the truncated protein upon differentiation, and that the contraction of the D4Z4 repeats abolishes this developmentally regulated process. It remains to be determined how D4Z4 contraction interferes with the normal induction of heterochromatin. Nevertheless, the use of FSHD iPS in this particular study, neatly illustrates the power of mutant pluripotent stem cells in resolving the molecular mechanisms contributing to disease pathogenesis.
During the past years it has become apparent that a third potential mechanism may act in the context of pathogenic repeat expansions. A novel form of translation was found by Ranum’s group, which results from translation in the absence of an initiating AUG codon next to disease causing unstable microsatellite repeats[95]. They found that the CAG repeat tract at the DMPK (DM1) and ATXN8 (SCA8) loci undergo noncanonical translation in disease relevant tissues of patients, resulting in the production of homopolymeric proteins in all three frames (CAG, AGC, and GCA). This newly discovered phenomenon was termed RAN translation, abbreviated for Repeat Associated Non-ATG translation.
To date, RAN translation has been shown to occur across four different types of repeat motifs: CAG, CTG, CGG and GGGGCC, in a growing number of conditions, including DM1, SCA8[95], FXTAS[96] and C9/ALS-FTD[39,40,50,97,98]. Among its effects, RAN translation leads to ubiquitin-positive intracellular protein inclusions, which may be toxic to cells[96]. Furthermore, researchers have shown that when repeats are bi-directionally transcribed to form sense and anti-sense transcripts, six potential homopolymeric proteins can be produced[48] as each transcript can be RAN translated in three different reading frames. Importantly, they provided evidence that long repeat tracts can express multiple RAN-translated products, simultaneously, in a single cell and suggested that RAN translation may contribute to disease pathogenesis by a protein gain-of-function mechanism[95]. In addition, as longer expansions are more liable to the accumulation of RAN translated products, it was speculated to be involved with disease anticipation[99]. Nonetheless, evidence for a role of RAN translation, if any, in disease pathogenesis still awaits further exploration.
Thus far, RAN translation has been documented in pluripotent stem cells only for C9/ALS-FTD. Indeed, postmitotic neurons derived from C9/ALS-FTD iPS cells have already been established, and were shown to present disease-relevant phenotypes that include RAN translation products in the form of cytoplasmic TDP-43 negative/p62 positive protein inclusions[51]. Immunofluorescent staining of the C9/ALS-FTD iPSNs (induced pluripotent stem cells derived neurons) revealed cytoplasmic poly-(Gly-Pro) inclusions similar to those observed in C9orf72 expansion carriers postmortem affected brains[53]. In addition, as the appearance of RAN peptide foci was coupled with increased sensitivity to cellular stress induced by autophagy inhibitors[51,53], it raised the possibility that this mechanism may contribute (directly or indirectly) to impaired autophagy function in ALS-FTD and possibly additional conditions associated with RAN translation[53].
There is by now enough evidence to indicate that mutant differentiated iPS cells can authentically recapitulate the late onset and tissue-specific cellular phenotypes that are typically seen in human C9orf72 postmortem brain samples. It should be interesting to explore whether similar cellular phenotypes can be reproduced in iPS cells derived from younger, yet asymptomatic, C9 mutation carriers. In addition, it would be useful to establish additional C9/ALS-FTD iPS cells from patients who carry small, intermediate and large expansions to correlate inclusions with repeat tract size, C9orf72 transcription and disease severity. Such associations are not yet fully understood. Nevertheless, it should be kept in mind that mitigation of RNA foci and RBP aggregation by antisense oligonucleotide (ASO) treatment in affected iPSNs had no effect on the formation of RAN-translation inclusions[53]. This would suggest that the detected homopolymeric inclusions may not have a major role in disease pathology. However, as only one type of di-peptide repeat was monitored [poly-(Gly-Pro)], it may be possible that in fact the other RAN peptides (such as the ones that are produced by antisense transcription) are crucial for the manifestation of the abovementioned cellular phenotypes. In any case, even if the RAN-translated products observed in iPS derived neurons will be proved to be irrelevant to disease pathology, they may be useful as a biomarker, providing an opportunity to develop a platform to test therapeutic agents for their effect on brain features of the disease. Compounds that will reverse the formation of the inclusions in the diseased cells are expected to offer a starting point for therapeutic development.
Many questions regarding RAN translation arise. For example, what is the clinical significance of RNA translation? Does RAN translation occur across all unstable microsatellite disorders? Is it age dependent? When and what determines its tissue specificity? In addition, it is still not yet clear which RNA structures and flanking sequences are required to facilitate RAN translation? Intrastrand RNA repeat associated hairpins and G-quadruplex structures have been suggested as candidates for RAN translation involvement[95,99-101]. When more stem cell based disease models (HESC and/or iPS) will become available, it is anticipated that human pluripotent stem cells will be important in clarifying at least some of these fundamental questions.
Repeat instability is a form of dynamic mutation that results from a change in the number of repeat units when ordered in tandem. Unlike static mutations, which involve permanent change in the DNA, repeat instabilities change across generations, and may vary between and within tissues of affected individuals (for comprehensive reviews see[102,103]). We refer to germ-line instability when the change in repeat number occurs during transmission from parent to offspring, and somatic instability when the change takes place in the somatic cells of the patient. Germ-line instability, which is frequently biased towards maternal or paternal transmissions, is typically associated with anticipation (reviewed in[103]). This is because the longer the repeat tract the more likely it will undergo expansion in the next generation. As a result, disease symptoms worsen from one generation to the next. However, in some pathologies repeat instability extends beyond the germ-line, leading to a high degree of variation in repeat tract length in affected individuals[14,104,105]. Unlike in germ-line repeat instability, the contribution of somatic instability to disease pathogenesis is not at all times clear. In some conditions there is an obvious correlation between expansion size and cell dysfunction[106,107], whereas in others this association in not noticeable[108,109]. Nevertheless, it is imperative to understand the molecular details of this mutation-associated phenomenon, considering that in some cases expansion size is clinically significant and may be used as a prognostic marker[110].
It is important to note that not all expansions are alike in range and location. While expansions residing in coding regions are usually restricted in length (tens of copies) and limited to germ-line mutations, noncoding expansions are frequently large (hundreds-thousands of copies) and typically coincide with somatic instability. Furthermore, in some disorders somatic instability can be restricted to early embryonic development or to highly replicating cells, whereas in others it may be tissue-specific or age-dependent, and may be limited to particular post-mitotic cell types. Therefore, while the type of mutation is common to all of these heritable and devastating conditions, the marked variation in the pattern and distribution of expansion size among them may stem from assorted mechanisms.
It is generally accepted that repeat instability results from local disruption of DNA replication, repair or recombination (for comprehensive review see[111]). Regardless of the mechanism, all proposed models for instability are based on the formation of unusual structures in the DNA (like hairpins and G-quadruplexes) that are eventually resolved by the addition (expansions) or deletion (contractions) of repetitive units[112-120]. It remains to be determined when and how these changes occur. In that sense, expansion carrying pluripotent stem cells provide an exceptional opportunity to gain more insights on the mechanisms that are involved with repeat instability. This is because they recapitulate early molecular events as they happen in vivo. They can be induced to differentiate into a wide range of cell types, allowing the monitoring of repeat instability in diseased relevant cells that are otherwise inaccessible for research like heart muscle cells or post-mitotic neurons. In addition, there is no evidence for further repeat amplification beyond the range observed in somatic cell of patients. Thus far, only a limited number of reports dealing with repeat instability in pluripotent cells have been published. All are descriptive in nature. For example, by following CTG expansion size at the DMPK gene of DM1-affected HESC lines, it was possible to demonstrate a change in repeat tract heterogeneity along with time in culture[121,122]. Furthermore, by inducing the cells to differentiate in vitro, instability was lost[122], apparently contradicting observations in vivo[123,124]. Nevertheless, it should be emphasized that tract-length heterogeneity within cell lines does not necessarily indicate that instability in fact exists, but rather proves that instability has occurred sometime before or during cell line growth in culture. This equally holds for CGG expansions at the FMR1 locus, where repeat length heterogeneity is clearly observed in a large cohort of FXS HESC lines[17,69]. Interestingly, in this case variations seem to be inversely correlated with CpG hypermethylation, a cis element that was formerly identified to constrain instability in FXS-affected fibroblasts[125]. Indeed, CGG somatic instability has been shown to be largely restricted to early fetal development and to cease with epigenetic gene silencing of FMR1[15]. In addition, as these cell lines are widely heterogeneous for expansion size and FMR1 hypermethylation, it may indicate that differentiation by itself is irrelevant to the repression of repeat instability in FXS, as formerly suggested[17]. It will be essential to generate single cell lineages from those FXS HESCs and monitor them for CGG repeat instability during growth in vitro, to unambiguously show that instability is, in fact, active in these cell cultures. Alternatively, it may be particularly useful to use patient iPS cells for these types of studies, considering the clonal origin of iPS cells. Two relevant examples for this application are C9/ALS-FTD iPS cells, where G4C2 tract size heterogeneity is evident both in undifferentiated and in in vitro-derived neurons[51], and the GAA expansion at the frataxin gene in FRDA[126,127]. On the other hand, the iPS based complimentary system should be cautiously tested before being adopted, as reprogramming may incorrectly enhance instability at repetitive loci as reported in FRDA iPS cells[128].
Apart from monitoring repeat instability along with time or as a consequence of reprogramming/differentiation in culture, diseased pluripotent stem cells (HESCs or iPS cells) may provide an unusual opportunity to gain new insights into the mechanisms underlying this mutational event. For example, it was shown that when the mismatch repair proteins, including MSH2 and MSH6, are downregulated upon differentiation repeat instability is dramatically enhanced in DM1 (CTG), HD (CAG)[122] and FRDA (GAA)[126,128] pluripotent stem cells, suggesting that repeat instability is assigned to difficulties in DNA repair at the these particular loci. This is in contrast to CGG somatic instability in FXS, where perturbations in DNA replication are assumed to be the underlying mechanism. In line with this perception is the study of Gerhardt and colleagues[129], who employed the DNA combing methodology to associate CGG expansion with alterations in origin of replication (ORI) usage and rate of progression in undifferentiated FXS HESCs. Using a pair of FXS HESC lines, these researchers reported a difference in ORI usage exclusive to undifferentiated FXS cells, proposing a switch in replication direction relative to the CGG repeats. According to their study, unlike in WT HESCs, replication proceeds predominantly from a downstream ORI in FXS HESCs, resulted in the replication of the CGGs by lagging strand synthesis (rather than by the leading strand) which favors expansions to occur. It remains to be determined whether the change in ORI usage in FXS HESCs is a cause or effect of the expansion, and whether it is mechanistically related to CGG somatic instability. The availability of a large cohort of FXS HESC lines that are diverse in their epigenetic state and CGG instability should help to address the aforementioned questions.
Lastly, it should mentioned that while all of the aforementioned studies relate to the issue of somatic instability using mutant pluripotent stem cells, it should be most attractive to utilize the same systems in order to recapitulate germ-line instability and better understand why such disease associated repetitive sequences become increasingly unstable during germ-line transmission. This can be accomplished once efficient protocols for inducing HESC/iPS to differentiate into primordial germ cells and fully matured gametes will be available. Indeed, differentiation of ES cells into fully matured oocytes[130] and the generation of viable offspring from these cells[131] have already been accomplished in mice, paving the way for such studies to be carried out in human cells in the near future.
HESCs are undifferentiated pluripotent cell lines with the potential for unrestricted self-renewal as well as the ability to differentiate into a wide range of cell types[132-134]. They are established from in vitro fertilization (IVF)-derived embryos, and can be obtained from genetically affected embryos following PGD procedures. The derivation of HESCs directly from diseased embryos provides a powerful tool for disease modeling[135,136] by affording the exploration of disease relevant tissues[137,138] and/or developmental stages[139] that are otherwise inaccessibly for research. The main limitation of this approach is that it relies on the availability of genetically affected embryos, a valuable resource that is accessible to only a small number of stem cell deriving centers worldwide. In such cases, a complement model system may be employed which relies on the derivation of patient-derived iPS cells; ES-like cells that are established by somatic cell reprogramming of patients’ cells in culture[140]. The primary advantage of this alternative system is that it facilitates the establishment of pluripotent cell lines that share many features with HESCs and harbor the disease causing mutation/s without the need for affected embryos. As such, and although artificially induced, it has become the most common alternative for modeling human diseases in culture by many researchers[16,141-146]. Nevertheless, iPS cells can be useful only if the disease is not embryonic lethal, and if complete erasure of the epigenetic modifications that are normally gained with cell differentiation do not interfere with investigation of disease pathogenesis. It should be mentioned that both HESC and iPS cells have been reported to carry typical chromosomal aberrations resulting from culture adaptation[147,148]. For this reason it is very important to perform routine characterization for chromosomal abnormalities in HESC and iPS cells lines.
The main benefits of both mutant HESC- and iPS-based model systems include the fact that they: (1) are human derived; (2) naturally carry disease causing mutations; (3) have the potential to differentiate into a wide range of cell types; (4) can self-renew without limitation while maintaining their undifferentiated and pluripotent potential; and (5) they mimic, to some extent, early embryo developmental stages as they spontaneously differentiate in vitro and in vivo. For all these reasons, they have become a commonly used model system for studying various disease associated mechanisms, particularly in cases where animal models do not authentically reproduce the affected human phenotype, or when the disease relevant cell types are unavailable for research.
In the context of disease causing unstable noncoding repeat expansions, which account for approximately half a dozen heritable conditions, the availability of mutant pluripotent stem cells is of particular importance. This is because most of these diseases are neuron-associated, restricting human-based research to a limited number of postmortem brain samples or to the use of disease irrelevant cell types. In addition, since noncoding expansions are hard to clone, it has been extremely difficult to artificially produce them in culture or to generate appropriate animal models[149-153]. In general, pluripotent stem cells seem to reproduce many of the pathological features that are observed in noncoding repeat expansion disorder patient cells[52,53,127,154-157]. Some of these features include nuclear RNA-protein foci, di-peptide cytoplasmic inclusions and hypermethylation. Therefore, stem cells are useful for basic research studies related to the different mechanisms that are involved with this group of disorders. By enlarging the number and repeat expansion heterogeneity of mutant stem cells cohort, we may gain new insights related to the underling mechanisms that are associated with these pathologies early during embryo development. Once efficient differentiation protocols will be established, it will also be possible to generate large amounts of cells from affected tissues in culture. Studying cell types that are particularly vulnerable in these diseases will improve our understanding regarding the tissue specificity seen in patients. Moreover, such cells can be utilized for drug screening and development studies. Stem cells provide a convenient platform to genetically intervene with the molecular defect that is inherently found in the cells. Considering the recent advancements in the field of artificially engineered nucleases (particularly the CRISPR/Cas9 system), it may be possible in the near future to insert, replace or remove repetitive DNA sequences from the genome of HESCs/iPS cells in a fairly uncomplicated procedure. This may hold a great promise for the development of new therapeutic approaches and hopefully will allow the ease, or even cure, pathologies resulting from unstable repeat expansions.
At present, mutant pluripotent stem cell lines are available for FXS, FXTAS, DM1, FRDA, C9/ALS-FTD, FSHD, and FRDA (Table 1). Most of these cell lines have been utilized for generating disease relevant cell types in order to validate the feasibility of the stem cell model system. In a few cases, where disease pathology is developmentally regulated, they have been particularly informative. For example, in FSHD stem cell lines facilitated the discovery of alternative polyadenylation signals responsible for the generation of toxic protein in a differentiation-dependent fashion[94]. In the case of FXS, an mRNA-mediated mechanism that drives epigenetic FMR1 gene silencing in affected neurons was uncovered in stem cells as well[73]. In other cases, particular cellular phenotypes in differentiated stem cell-derived cultures have been monitored by effect of therapeutic agents or by defining proteins that are associated with pathological protein inclusions, as reported in C9/ALS iPS-derived neuronal cells[52,53].
| Disease | Molecular mechanism | Ref. |
| DM1 | Toxic RNA gain-of-function | [45,46] |
| C9/ALS-FTD | [47-49,51-53] | |
| FXS | Reaped-mediated epigenetic modifications | [17,62,69,71-76,78] |
| C9/ALS-FTD | [52] | |
| FSHD | [92] | |
| C9/ALS-FTD | RAN translation | [51,53] |
| FXS | Repeat instability | [17,69,127] |
| C9/ALS-FTD | [51] | |
| FDRA | [124-126] | |
| DM1 | [119,120] |
As a note of caution, it should be understood that the development of efficient stem cell differentiation protocols is not that straightforward. In particular, it can be quite challenging to use stem cells for the derivation of homogeneous cell populations that are functionally impaired. Moreover, molecular phenotypes in mutant pluripotent cells must be validated to see if they reproduce molecular events that are observed in diseased individuals. For example, despite all the benefits of iPS cells, investigators are becoming increasingly more aware of epigenetic instability in these cell types[158-161] as many iPSs wrongly acquire hypermethylation as a consequence of reprogramming[78,162-164]. Accordingly, a comparison between mutant HESCs and patient-derived iPS cells will be essential in future investigations of the epigenetic aspects of repeat expansion diseases especially because many mechanistic questions still remain to be addressed in this area. Unanswered questions in the field include what molecular events lead to hypermethylation of repeat expansions? What pathogenic role do nuclear riboprotein or cytoplasmic di-pepetide inclusions have, and what are the proteins that are sequestered into these structures? What are the mechanisms that are responsible for germ-line and somatic repeat instability, and why does somatic instability differ according to patient age and by tissue-specificity among differing genomic loci? Given proper experimental conditions, the answers to these riveting questions may be waiting to be uncovered by stem cell-based model systems.
P- Reviewer: Cho SG, Fukuda S, Kawasaki H, Majumdar APN, Zhou FC S- Editor: Ji FF L- Editor: A E- Editor: Wang CH
| 1. | Gatchel JR, Zoghbi HY. Diseases of unstable repeat expansion: mechanisms and common principles. Nat Rev Genet. 2005;6:743-755. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 561] [Cited by in F6Publishing: 556] [Article Influence: 29.3] [Reference Citation Analysis (0)] |
| 2. | Dick KA, Margolis JM, Day JW, Ranum LP. Dominant non-coding repeat expansions in human disease. Genome Dyn. 2006;1:67-83. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 18] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 3. | Hagerman RJ, Leehey M, Heinrichs W, Tassone F, Wilson R, Hills J, Grigsby J, Gage B, Hagerman PJ. Intention tremor, parkinsonism, and generalized brain atrophy in male carriers of fragile X. Neurology. 2001;57:127-130. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 664] [Cited by in F6Publishing: 653] [Article Influence: 28.4] [Reference Citation Analysis (0)] |
| 4. | Oberlé I, Rousseau F, Heitz D, Kretz C, Devys D, Hanauer A, Boué J, Bertheas MF, Mandel JL. Instability of a 550-base pair DNA segment and abnormal methylation in fragile X syndrome. Science. 1991;252:1097-1102. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1044] [Cited by in F6Publishing: 1004] [Article Influence: 30.4] [Reference Citation Analysis (0)] |
| 5. | Verkerk AJ, Pieretti M, Sutcliffe JS, Fu YH, Kuhl DP, Pizzuti A, Reiner O, Richards S, Victoria MF, Zhang FP. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991;65:905-914. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2433] [Cited by in F6Publishing: 2396] [Article Influence: 72.6] [Reference Citation Analysis (0)] |
| 6. | Brook JD, McCurrach ME, Harley HG, Buckler AJ, Church D, Aburatani H, Hunter K, Stanton VP, Thirion JP, Hudson T. Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3’ end of a transcript encoding a protein kinase family member. Cell. 1992;69:385. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1702] [Cited by in F6Publishing: 1630] [Article Influence: 50.9] [Reference Citation Analysis (0)] |
| 7. | Fu YH, Pizzuti A, Fenwick RG, King J, Rajnarayan S, Dunne PW, Dubel J, Nasser GA, Ashizawa T, de Jong P. An unstable triplet repeat in a gene related to myotonic muscular dystrophy. Science. 1992;255:1256-1258. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1043] [Cited by in F6Publishing: 1001] [Article Influence: 31.3] [Reference Citation Analysis (0)] |
| 8. | Mahadevan M, Tsilfidis C, Sabourin L, Shutler G, Amemiya C, Jansen G, Neville C, Narang M, Barceló J, O’Hoy K. Myotonic dystrophy mutation: an unstable CTG repeat in the 3’ untranslated region of the gene. Science. 1992;255:1253-1255. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1148] [Cited by in F6Publishing: 1126] [Article Influence: 35.2] [Reference Citation Analysis (0)] |
| 9. | Liquori CL, Ricker K, Moseley ML, Jacobsen JF, Kress W, Naylor SL, Day JW, Ranum LP. Myotonic dystrophy type 2 caused by a CCTG expansion in intron 1 of ZNF9. Science. 2001;293:864-867. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 913] [Cited by in F6Publishing: 834] [Article Influence: 36.3] [Reference Citation Analysis (0)] |
| 10. | Campuzano V, Montermini L, Moltò MD, Pianese L, Cossée M, Cavalcanti F, Monros E, Rodius F, Duclos F, Monticelli A. Friedreich’s ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science. 1996;271:1423-1427. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2041] [Cited by in F6Publishing: 1876] [Article Influence: 67.0] [Reference Citation Analysis (0)] |
| 11. | Xi Z, Zinman L, Moreno D, Schymick J, Liang Y, Sato C, Zheng Y, Ghani M, Dib S, Keith J. Hypermethylation of the CpG island near the G4C2 repeat in ALS with a C9orf72 expansion. Am J Hum Genet. 2013;92:981-989. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 204] [Cited by in F6Publishing: 205] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 12. | Murray ME, DeJesus-Hernandez M, Rutherford NJ, Baker M, Duara R, Graff-Radford NR, Wszolek ZK, Ferman TJ, Josephs KA, Boylan KB. Clinical and neuropathologic heterogeneity of c9FTD/ALS associated with hexanucleotide repeat expansion in C9ORF72. Acta Neuropathol. 2011;122:673-690. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 224] [Cited by in F6Publishing: 237] [Article Influence: 18.2] [Reference Citation Analysis (0)] |
| 13. | Nelson DL, Orr HT, Warren ST. The unstable repeats--three evolving faces of neurological disease. Neuron. 2013;77:825-843. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 166] [Cited by in F6Publishing: 148] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 14. | Fu YH, Kuhl DP, Pizzuti A, Pieretti M, Sutcliffe JS, Richards S, Verkerk AJ, Holden JJ, Fenwick RG, Warren ST. Variation of the CGG repeat at the fragile X site results in genetic instability: resolution of the Sherman paradox. Cell. 1991;67:1047-1058. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1441] [Cited by in F6Publishing: 1403] [Article Influence: 42.5] [Reference Citation Analysis (0)] |
| 15. | Wöhrle D, Hennig I, Vogel W, Steinbach P. Mitotic stability of fragile X mutations in differentiated cells indicates early post-conceptional trinucleotide repeat expansion. Nat Genet. 1993;4:140-142. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 129] [Cited by in F6Publishing: 135] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 16. | Park IH, Arora N, Huo H, Maherali N, Ahfeldt T, Shimamura A, Lensch MW, Cowan C, Hochedlinger K, Daley GQ. Disease-specific induced pluripotent stem cells. Cell. 2008;134:877-886. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1655] [Cited by in F6Publishing: 1577] [Article Influence: 98.6] [Reference Citation Analysis (0)] |
| 17. | Eiges R, Urbach A, Malcov M, Frumkin T, Schwartz T, Amit A, Yaron Y, Eden A, Yanuka O, Benvenisty N. Developmental study of fragile X syndrome using human embryonic stem cells derived from preimplantation genetically diagnosed embryos. Cell Stem Cell. 2007;1:568-577. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 226] [Cited by in F6Publishing: 208] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 18. | Dimos JT, Rodolfa KT, Niakan KK, Weisenthal LM, Mitsumoto H, Chung W, Croft GF, Saphier G, Leibel R, Goland R. Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science. 2008;321:1218-1221. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1445] [Cited by in F6Publishing: 1379] [Article Influence: 86.2] [Reference Citation Analysis (0)] |
| 19. | An MC, Zhang N, Scott G, Montoro D, Wittkop T, Mooney S, Melov S, Ellerby LM. Genetic correction of Huntington’s disease phenotypes in induced pluripotent stem cells. Cell Stem Cell. 2012;11:253-263. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 266] [Cited by in F6Publishing: 261] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 20. | Bard J, Wall MD, Lazari O, Arjomand J, Munoz-Sanjuan I. Advances in huntington disease drug discovery: novel approaches to model disease phenotypes. J Biomol Screen. 2014;19:191-204. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 12] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 21. | Kaye JA, Finkbeiner S. Modeling Huntington’s disease with induced pluripotent stem cells. Mol Cell Neurosci. 2013;56:50-64. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 51] [Cited by in F6Publishing: 46] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 22. | Li M, Suzuki K, Kim NY, Liu GH, Izpisua Belmonte JC. A cut above the rest: targeted genome editing technologies in human pluripotent stem cells. J Biol Chem. 2014;289:4594-4599. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 91] [Cited by in F6Publishing: 96] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 23. | Taneja KL, McCurrach M, Schalling M, Housman D, Singer RH. Foci of trinucleotide repeat transcripts in nuclei of myotonic dystrophy cells and tissues. J Cell Biol. 1995;128:995-1002. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 447] [Cited by in F6Publishing: 469] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 24. | Margolis JM, Schoser BG, Moseley ML, Day JW, Ranum LP. DM2 intronic expansions: evidence for CCUG accumulation without flanking sequence or effects on ZNF9 mRNA processing or protein expression. Hum Mol Genet. 2006;15:1808-1815. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 84] [Cited by in F6Publishing: 92] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 25. | Sellier C, Rau F, Liu Y, Tassone F, Hukema RK, Gattoni R, Schneider A, Richard S, Willemsen R, Elliott DJ. Sam68 sequestration and partial loss of function are associated with splicing alterations in FXTAS patients. EMBO J. 2010;29:1248-1261. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 269] [Cited by in F6Publishing: 282] [Article Influence: 20.1] [Reference Citation Analysis (0)] |
| 26. | DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Flynn H, Adamson J. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245-256. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3328] [Cited by in F6Publishing: 3520] [Article Influence: 270.8] [Reference Citation Analysis (0)] |
| 27. | Li LB, Yu Z, Teng X, Bonini NM. RNA toxicity is a component of ataxin-3 degeneration in Drosophila. Nature. 2008;453:1107-1111. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 237] [Cited by in F6Publishing: 234] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 28. | Daughters RS, Tuttle DL, Gao W, Ikeda Y, Moseley ML, Ebner TJ, Swanson MS, Ranum LP. RNA gain-of-function in spinocerebellar ataxia type 8. PLoS Genet. 2009;5:e1000600. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 218] [Cited by in F6Publishing: 226] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 29. | White MC, Gao R, Xu W, Mandal SM, Lim JG, Hazra TK, Wakamiya M, Edwards SF, Raskin S, Teive HA. Inactivation of hnRNP K by expanded intronic AUUCU repeat induces apoptosis via translocation of PKCdelta to mitochondria in spinocerebellar ataxia 10. PLoS Genet. 2010;6:e1000984. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 92] [Cited by in F6Publishing: 95] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 30. | Sato N, Amino T, Kobayashi K, Asakawa S, Ishiguro T, Tsunemi T, Takahashi M, Matsuura T, Flanigan KM, Iwasaki S. Spinocerebellar ataxia type 31 is associated with “inserted” penta-nucleotide repeats containing (TGGAA)n. Am J Hum Genet. 2009;85:544-557. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 206] [Cited by in F6Publishing: 203] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 31. | Rudnicki DD, Holmes SE, Lin MW, Thornton CA, Ross CA, Margolis RL. Huntington’s disease--like 2 is associated with CUG repeat-containing RNA foci. Ann Neurol. 2007;61:272-282. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 118] [Cited by in F6Publishing: 115] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 32. | Miller JW, Urbinati CR, Teng-Umnuay P, Stenberg MG, Byrne BJ, Thornton CA, Swanson MS. Recruitment of human muscleblind proteins to (CUG)(n) expansions associated with myotonic dystrophy. EMBO J. 2000;19:4439-4448. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 684] [Cited by in F6Publishing: 679] [Article Influence: 28.3] [Reference Citation Analysis (0)] |
| 33. | Timchenko NA, Cai ZJ, Welm AL, Reddy S, Ashizawa T, Timchenko LT. RNA CUG repeats sequester CUGBP1 and alter protein levels and activity of CUGBP1. J Biol Chem. 2001;276:7820-7826. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 241] [Cited by in F6Publishing: 248] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 34. | Botta A, Rinaldi F, Catalli C, Vergani L, Bonifazi E, Romeo V, Loro E, Viola A, Angelini C, Novelli G. The CTG repeat expansion size correlates with the splicing defects observed in muscles from myotonic dystrophy type 1 patients. J Med Genet. 2008;45:639-646. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 40] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 35. | Du H, Cline MS, Osborne RJ, Tuttle DL, Clark TA, Donohue JP, Hall MP, Shiue L, Swanson MS, Thornton CA. Aberrant alternative splicing and extracellular matrix gene expression in mouse models of myotonic dystrophy. Nat Struct Mol Biol. 2010;17:187-193. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 256] [Cited by in F6Publishing: 250] [Article Influence: 17.9] [Reference Citation Analysis (0)] |
| 36. | Kanadia RN, Johnstone KA, Mankodi A, Lungu C, Thornton CA, Esson D, Timmers AM, Hauswirth WW, Swanson MS. A muscleblind knockout model for myotonic dystrophy. Science. 2003;302:1978-1980. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 560] [Cited by in F6Publishing: 546] [Article Influence: 26.0] [Reference Citation Analysis (0)] |
| 37. | Tassone F, Iwahashi C, Hagerman PJ. FMR1 RNA within the intranuclear inclusions of fragile X-associated tremor/ataxia syndrome (FXTAS). RNA Biol. 2004;1:103-105. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 195] [Cited by in F6Publishing: 190] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 38. | Ladd PD, Smith LE, Rabaia NA, Moore JM, Georges SA, Hansen RS, Hagerman RJ, Tassone F, Tapscott SJ, Filippova GN. An antisense transcript spanning the CGG repeat region of FMR1 is upregulated in premutation carriers but silenced in full mutation individuals. Hum Mol Genet. 2007;16:3174-3187. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 200] [Cited by in F6Publishing: 213] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 39. | Mori K, Weng SM, Arzberger T, May S, Rentzsch K, Kremmer E, Schmid B, Kretzschmar HA, Cruts M, Van Broeckhoven C. The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science. 2013;339:1335-1338. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 898] [Cited by in F6Publishing: 939] [Article Influence: 85.4] [Reference Citation Analysis (0)] |
| 40. | Zu T, Liu Y, Bañez-Coronel M, Reid T, Pletnikova O, Lewis J, Miller TM, Harms MB, Falchook AE, Subramony SH. RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proc Natl Acad Sci USA. 2013;110:E4968-E4977. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 562] [Cited by in F6Publishing: 585] [Article Influence: 53.2] [Reference Citation Analysis (0)] |
| 41. | de Haro M, Al-Ramahi I, De Gouyon B, Ukani L, Rosa A, Faustino NA, Ashizawa T, Cooper TA, Botas J. MBNL1 and CUGBP1 modify expanded CUG-induced toxicity in a Drosophila model of myotonic dystrophy type 1. Hum Mol Genet. 2006;15:2138-2145. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 109] [Cited by in F6Publishing: 116] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 42. | deLorimier E, Coonrod LA, Copperman J, Taber A, Reister EE, Sharma K, Todd PK, Guenza MG, Berglund JA. Modifications to toxic CUG RNAs induce structural stability, rescue mis-splicing in a myotonic dystrophy cell model and reduce toxicity in a myotonic dystrophy zebrafish model. Nucleic Acids Res. 2014;42:12768-12778. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 23] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 43. | Jin P, Duan R, Qurashi A, Qin Y, Tian D, Rosser TC, Liu H, Feng Y, Warren ST. Pur alpha binds to rCGG repeats and modulates repeat-mediated neurodegeneration in a Drosophila model of fragile X tremor/ataxia syndrome. Neuron. 2007;55:556-564. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 240] [Cited by in F6Publishing: 240] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 44. | Kanadia RN, Shin J, Yuan Y, Beattie SG, Wheeler TM, Thornton CA, Swanson MS. Reversal of RNA missplicing and myotonia after muscleblind overexpression in a mouse poly(CUG) model for myotonic dystrophy. Proc Natl Acad Sci USA. 2006;103:11748-11753. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 287] [Cited by in F6Publishing: 282] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 45. | Denis JA, Gauthier M, Rachdi L, Aubert S, Giraud-Triboult K, Poydenot P, Benchoua A, Champon B, Maury Y, Baldeschi C. mTOR-dependent proliferation defect in human ES-derived neural stem cells affected by myotonic dystrophy type 1. J Cell Sci. 2013;126:1763-1772. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 33] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 46. | Xia G, Santostefano KE, Goodwin M, Liu J, Subramony SH, Swanson MS, Terada N, Ashizawa T. Generation of neural cells from DM1 induced pluripotent stem cells as cellular model for the study of central nervous system neuropathogenesis. Cell Reprogram. 2013;15:166-177. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 41] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 47. | Morita M, Al-Chalabi A, Andersen PM, Hosler B, Sapp P, Englund E, Mitchell JE, Habgood JJ, de Belleroche J, Xi J. A locus on chromosome 9p confers susceptibility to ALS and frontotemporal dementia. Neurology. 2006;66:839-844. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 259] [Cited by in F6Publishing: 260] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 48. | Pearson JP, Williams NM, Majounie E, Waite A, Stott J, Newsway V, Murray A, Hernandez D, Guerreiro R, Singleton AB. Familial frontotemporal dementia with amyotrophic lateral sclerosis and a shared haplotype on chromosome 9p. J Neurol. 2011;258:647-655. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 60] [Cited by in F6Publishing: 62] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 49. | Vance C, Al-Chalabi A, Ruddy D, Smith BN, Hu X, Sreedharan J, Siddique T, Schelhaas HJ, Kusters B, Troost D. Familial amyotrophic lateral sclerosis with frontotemporal dementia is linked to a locus on chromosome 9p13.2-21.3. Brain. 2006;129:868-876. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 281] [Cited by in F6Publishing: 280] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 50. | Gendron TF, Bieniek KF, Zhang YJ, Jansen-West K, Ash PE, Caulfield T, Daughrity L, Dunmore JH, Castanedes-Casey M, Chew J. Antisense transcripts of the expanded C9ORF72 hexanucleotide repeat form nuclear RNA foci and undergo repeat-associated non-ATG translation in c9FTD/ALS. Acta Neuropathol. 2013;126:829-844. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 415] [Cited by in F6Publishing: 436] [Article Influence: 39.6] [Reference Citation Analysis (0)] |
| 51. | Almeida S, Gascon E, Tran H, Chou HJ, Gendron TF, Degroot S, Tapper AR, Sellier C, Charlet-Berguerand N, Karydas A. Modeling key pathological features of frontotemporal dementia with C9ORF72 repeat expansion in iPSC-derived human neurons. Acta Neuropathol. 2013;126:385-399. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 257] [Cited by in F6Publishing: 232] [Article Influence: 21.1] [Reference Citation Analysis (0)] |
| 52. | Sareen D, O’Rourke JG, Meera P, Muhammad AK, Grant S, Simpkinson M, Bell S, Carmona S, Ornelas L, Sahabian A. Targeting RNA foci in iPSC-derived motor neurons from ALS patients with a C9ORF72 repeat expansion. Sci Transl Med. 2013;5:208ra149. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 501] [Cited by in F6Publishing: 507] [Article Influence: 50.7] [Reference Citation Analysis (0)] |
| 53. | Donnelly CJ, Zhang PW, Pham JT, Haeusler AR, Mistry NA, Vidensky S, Daley EL, Poth EM, Hoover B, Fines DM. RNA toxicity from the ALS/FTD C9ORF72 expansion is mitigated by antisense intervention. Neuron. 2013;80:415-428. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 661] [Cited by in F6Publishing: 682] [Article Influence: 68.2] [Reference Citation Analysis (0)] |
| 54. | Mankodi A, Logigian E, Callahan L, McClain C, White R, Henderson D, Krym M, Thornton CA. Myotonic dystrophy in transgenic mice expressing an expanded CUG repeat. Science. 2000;289:1769-1773. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 551] [Cited by in F6Publishing: 519] [Article Influence: 21.6] [Reference Citation Analysis (0)] |
| 55. | Xu Z, Poidevin M, Li X, Li Y, Shu L, Nelson DL, Li H, Hales CM, Gearing M, Wingo TS. Expanded GGGGCC repeat RNA associated with amyotrophic lateral sclerosis and frontotemporal dementia causes neurodegeneration. Proc Natl Acad Sci USA. 2013;110:7778-7783. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 260] [Cited by in F6Publishing: 260] [Article Influence: 23.6] [Reference Citation Analysis (0)] |
| 56. | Coppola G, Choi SH, Santos MM, Miranda CJ, Tentler D, Wexler EM, Pandolfo M, Geschwind DH. Gene expression profiling in frataxin deficient mice: microarray evidence for significant expression changes without detectable neurodegeneration. Neurobiol Dis. 2006;22:302-311. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 46] [Cited by in F6Publishing: 47] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 57. | Dansithong W, Wolf CM, Sarkar P, Paul S, Chiang A, Holt I, Morris GE, Branco D, Sherwood MC, Comai L. Cytoplasmic CUG RNA foci are insufficient to elicit key DM1 features. PLoS One. 2008;3:e3968. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 34] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 58. | den Broeder MJ, van der Linde H, Brouwer JR, Oostra BA, Willemsen R, Ketting RF. Generation and characterization of FMR1 knockout zebrafish. PLoS One. 2009;4:e7910. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 70] [Cited by in F6Publishing: 75] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 59. | Martelli A, Friedman LS, Reutenauer L, Messaddeq N, Perlman SL, Lynch DR, Fedosov K, Schulz JB, Pandolfo M, Puccio H. Clinical data and characterization of the liver conditional mouse model exclude neoplasia as a non-neurological manifestation associated with Friedreich’s ataxia. Dis Model Mech. 2012;5:860-869. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 31] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 60. | Santos MM, Miranda CJ, Levy JE, Montross LK, Cossée M, Sequeiros J, Andrews N, Koenig M, Pandolfo M. Iron metabolism in mice with partial frataxin deficiency. Cerebellum. 2003;2:146-153. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 5] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 61. | Wheeler TM, Krym MC, Thornton CA. Ribonuclear foci at the neuromuscular junction in myotonic dystrophy type 1. Neuromuscul Disord. 2007;17:242-247. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 39] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 62. | Sutcliffe JS, Nelson DL, Zhang F, Pieretti M, Caskey CT, Saxe D, Warren ST. DNA methylation represses FMR-1 transcription in fragile X syndrome. Hum Mol Genet. 1992;1:397-400. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 493] [Cited by in F6Publishing: 478] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 63. | Xi Z, Rainero I, Rubino E, Pinessi L, Bruni AC, Maletta RG, Nacmias B, Sorbi S, Galimberti D, Surace EI. Hypermethylation of the CpG-island near the C9orf72 G4C2-repeat expansion in FTLD patients. Hum Mol Genet. 2014;23:5630-5637. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 62] [Cited by in F6Publishing: 65] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 64. | Klesert TR, Otten AD, Bird TD, Tapscott SJ. Trinucleotide repeat expansion at the myotonic dystrophy locus reduces expression of DMAHP. Nat Genet. 1997;16:402-406. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 183] [Cited by in F6Publishing: 191] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 65. | Kumari D, Biacsi RE, Usdin K. Repeat expansion affects both transcription initiation and elongation in friedreich ataxia cells. J Biol Chem. 2011;286:4209-4215. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 76] [Cited by in F6Publishing: 70] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 66. | Lemmers RJ, van der Vliet PJ, Klooster R, Sacconi S, Camaño P, Dauwerse JG, Snider L, Straasheijm KR, van Ommen GJ, Padberg GW. A unifying genetic model for facioscapulohumeral muscular dystrophy. Science. 2010;329:1650-1653. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 565] [Cited by in F6Publishing: 530] [Article Influence: 37.9] [Reference Citation Analysis (0)] |
| 67. | Godler DE, Slater HR, Bui QM, Ono M, Gehling F, Francis D, Amor DJ, Hopper JL, Hagerman R, Loesch DZ. FMR1 intron 1 methylation predicts FMRP expression in blood of female carriers of expanded FMR1 alleles. J Mol Diagn. 2011;13:528-536. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 28] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 68. | López Castel A, Nakamori M, Tomé S, Chitayat D, Gourdon G, Thornton CA, Pearson CE. Expanded CTG repeat demarcates a boundary for abnormal CpG methylation in myotonic dystrophy patient tissues. Hum Mol Genet. 2011;20:1-15. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 104] [Cited by in F6Publishing: 99] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 69. | Avitzour M, Mor-Shaked H, Yanovsky-Dagan S, Aharoni S, Altarescu G, Renbaum P, Eldar-Geva T, Schonberger O, Levy-Lahad E, Epsztejn-Litman S. FMR1 epigenetic silencing commonly occurs in undifferentiated fragile X-affected embryonic stem cells. Stem Cell Reports. 2014;3:699-706. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 54] [Cited by in F6Publishing: 51] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 70. | Bell AC, West AG, Felsenfeld G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 1999;98:387-396. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 814] [Cited by in F6Publishing: 814] [Article Influence: 32.6] [Reference Citation Analysis (0)] |
| 71. | Hark AT, Schoenherr CJ, Katz DJ, Ingram RS, Levorse JM, Tilghman SM. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000;405:486-489. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1117] [Cited by in F6Publishing: 1097] [Article Influence: 45.7] [Reference Citation Analysis (0)] |
| 72. | Lanni S, Goracci M, Borrelli L, Mancano G, Chiurazzi P, Moscato U, Ferrè F, Helmer-Citterich M, Tabolacci E, Neri G. Role of CTCF protein in regulating FMR1 locus transcription. PLoS Genet. 2013;9:e1003601. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 34] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 73. | Colak D, Zaninovic N, Cohen MS, Rosenwaks Z, Yang WY, Gerhardt J, Disney MD, Jaffrey SR. Promoter-bound trinucleotide repeat mRNA drives epigenetic silencing in fragile X syndrome. Science. 2014;343:1002-1005. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 218] [Cited by in F6Publishing: 219] [Article Influence: 21.9] [Reference Citation Analysis (0)] |
| 74. | Burman RW, Popovich BW, Jacky PB, Turker MS. Fully expanded FMR1 CGG repeats exhibit a length- and differentiation-dependent instability in cell hybrids that is independent of DNA methylation. Hum Mol Genet. 1999;8:2293-2302. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 30] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 75. | Alisch RS, Wang T, Chopra P, Visootsak J, Conneely KN, Warren ST. Genome-wide analysis validates aberrant methylation in fragile X syndrome is specific to the FMR1 locus. BMC Med Genet. 2013;14:18. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 39] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 76. | Sheridan SD, Theriault KM, Reis SA, Zhou F, Madison JM, Daheron L, Loring JF, Haggarty SJ. Epigenetic characterization of the FMR1 gene and aberrant neurodevelopment in human induced pluripotent stem cell models of fragile X syndrome. PLoS One. 2011;6:e26203. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 233] [Cited by in F6Publishing: 227] [Article Influence: 17.5] [Reference Citation Analysis (0)] |
| 77. | Urbach A, Bar-Nur O, Daley GQ, Benvenisty N. Differential modeling of fragile X syndrome by human embryonic stem cells and induced pluripotent stem cells. Cell Stem Cell. 2010;6:407-411. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 313] [Cited by in F6Publishing: 291] [Article Influence: 20.8] [Reference Citation Analysis (0)] |
| 78. | de Esch CE, Ghazvini M, Loos F, Schelling-Kazaryan N, Widagdo W, Munshi ST, van der Wal E, Douben H, Gunhanlar N, Kushner SA. Epigenetic characterization of the FMR1 promoter in induced pluripotent stem cells from human fibroblasts carrying an unmethylated full mutation. Stem Cell Reports. 2014;3:548-555. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 42] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 79. | Planello AC, Ji J, Sharma V, Singhania R, Mbabaali F, Müller F, Alfaro JA, Bock C, De Carvalho DD, Batada NN. Aberrant DNA methylation reprogramming during induced pluripotent stem cell generation is dependent on the choice of reprogramming factors. Cell Regen (Lond). 2014;3:4. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 18] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 80. | Bar-Nur O, Caspi I, Benvenisty N. Molecular analysis of FMR1 reactivation in fragile-X induced pluripotent stem cells and their neuronal derivatives. J Mol Cell Biol. 2012;4:180-183. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 65] [Cited by in F6Publishing: 68] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 81. | Fratta P, Poulter M, Lashley T, Rohrer JD, Polke JM, Beck J, Ryan N, Hensman D, Mizielinska S, Waite AJ. Homozygosity for the C9orf72 GGGGCC repeat expansion in frontotemporal dementia. Acta Neuropathol. 2013;126:401-409. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 119] [Cited by in F6Publishing: 111] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 82. | Belzil VV, Bauer PO, Prudencio M, Gendron TF, Stetler CT, Yan IK, Pregent L, Daughrity L, Baker MC, Rademakers R. Reduced C9orf72 gene expression in c9FTD/ALS is caused by histone trimethylation, an epigenetic event detectable in blood. Acta Neuropathol. 2013;126:895-905. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 221] [Cited by in F6Publishing: 227] [Article Influence: 20.6] [Reference Citation Analysis (0)] |
| 83. | Russ J, Liu EY, Wu K, Neal D, Suh E, Irwin DJ, McMillan CT, Harms MB, Cairns NJ, Wood EM. Hypermethylation of repeat expanded C9orf72 is a clinical and molecular disease modifier. Acta Neuropathol. 2015;129:39-52. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 90] [Cited by in F6Publishing: 84] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 84. | Ciura S, Lattante S, Le Ber I, Latouche M, Tostivint H, Brice A, Kabashi E. Loss of function of C9orf72 causes motor deficits in a zebrafish model of amyotrophic lateral sclerosis. Ann Neurol. 2013;74:180-187. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 94] [Cited by in F6Publishing: 187] [Article Influence: 18.7] [Reference Citation Analysis (0)] |
| 85. | Lagier-Tourenne C, Baughn M, Rigo F, Sun S, Liu P, Li HR, Jiang J, Watt AT, Chun S, Katz M. Targeted degradation of sense and antisense C9orf72 RNA foci as therapy for ALS and frontotemporal degeneration. Proc Natl Acad Sci USA. 2013;110:E4530-E4539. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 420] [Cited by in F6Publishing: 435] [Article Influence: 39.5] [Reference Citation Analysis (0)] |
| 86. | Liu EY, Russ J, Wu K, Neal D, Suh E, McNally AG, Irwin DJ, Van Deerlin VM, Lee EB. C9orf72 hypermethylation protects against repeat expansion-associated pathology in ALS/FTD. Acta Neuropathol. 2014;128:525-541. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 130] [Cited by in F6Publishing: 132] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 87. | Tupler R, Berardinelli A, Barbierato L, Frants R, Hewitt JE, Lanzi G, Maraschio P, Tiepolo L. Monosomy of distal 4q does not cause facioscapulohumeral muscular dystrophy. J Med Genet. 1996;33:366-370. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 104] [Cited by in F6Publishing: 92] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 88. | van Deutekom JC, Wijmenga C, van Tienhoven EA, Gruter AM, Hewitt JE, Padberg GW, van Ommen GJ, Hofker MH, Frants RR. FSHD associated DNA rearrangements are due to deletions of integral copies of a 3.2 kb tandemly repeated unit. Hum Mol Genet. 1993;2:2037-2042. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 390] [Cited by in F6Publishing: 379] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 89. | Wijmenga C, Hewitt JE, Sandkuijl LA, Clark LN, Wright TJ, Dauwerse HG, Gruter AM, Hofker MH, Moerer P, Williamson R. Chromosome 4q DNA rearrangements associated with facioscapulohumeral muscular dystrophy. Nat Genet. 1992;2:26-30. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 482] [Cited by in F6Publishing: 468] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 90. | Dixit M, Ansseau E, Tassin A, Winokur S, Shi R, Qian H, Sauvage S, Mattéotti C, van Acker AM, Leo O. DUX4, a candidate gene of facioscapulohumeral muscular dystrophy, encodes a transcriptional activator of PITX1. Proc Natl Acad Sci USA. 2007;104:18157-18162. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 272] [Cited by in F6Publishing: 276] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 91. | Kowaljow V, Marcowycz A, Ansseau E, Conde CB, Sauvage S, Mattéotti C, Arias C, Corona ED, Nuñez NG, Leo O. The DUX4 gene at the FSHD1A locus encodes a pro-apoptotic protein. Neuromuscul Disord. 2007;17:611-623. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 245] [Cited by in F6Publishing: 246] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 92. | van Overveld PG, Lemmers RJ, Sandkuijl LA, Enthoven L, Winokur ST, Bakels F, Padberg GW, van Ommen GJ, Frants RR, van der Maarel SM. Hypomethylation of D4Z4 in 4q-linked and non-4q-linked facioscapulohumeral muscular dystrophy. Nat Genet. 2003;35:315-317. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 292] [Cited by in F6Publishing: 304] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 93. | Zeng W, de Greef JC, Chen YY, Chien R, Kong X, Gregson HC, Winokur ST, Pyle A, Robertson KD, Schmiesing JA. Specific loss of histone H3 lysine 9 trimethylation and HP1gamma/cohesin binding at D4Z4 repeats is associated with facioscapulohumeral dystrophy (FSHD). PLoS Genet. 2009;5:e1000559. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 215] [Cited by in F6Publishing: 218] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 94. | Snider L, Geng LN, Lemmers RJ, Kyba M, Ware CB, Nelson AM, Tawil R, Filippova GN, van der Maarel SM, Tapscott SJ. Facioscapulohumeral dystrophy: incomplete suppression of a retrotransposed gene. PLoS Genet. 2010;6:e1001181. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 361] [Cited by in F6Publishing: 350] [Article Influence: 25.0] [Reference Citation Analysis (0)] |
| 95. | Zu T, Gibbens B, Doty NS, Gomes-Pereira M, Huguet A, Stone MD, Margolis J, Peterson M, Markowski TW, Ingram MA. Non-ATG-initiated translation directed by microsatellite expansions. Proc Natl Acad Sci USA. 2011;108:260-265. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 639] [Cited by in F6Publishing: 645] [Article Influence: 46.1] [Reference Citation Analysis (0)] |
| 96. | Todd PK, Oh SY, Krans A, He F, Sellier C, Frazer M, Renoux AJ, Chen KC, Scaglione KM, Basrur V. CGG repeat-associated translation mediates neurodegeneration in fragile X tremor ataxia syndrome. Neuron. 2013;78:440-455. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 336] [Cited by in F6Publishing: 347] [Article Influence: 31.5] [Reference Citation Analysis (0)] |
| 97. | Ash PE, Bieniek KF, Gendron TF, Caulfield T, Lin WL, Dejesus-Hernandez M, van Blitterswijk MM, Jansen-West K, Paul JW, Rademakers R. Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron. 2013;77:639-646. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 804] [Cited by in F6Publishing: 839] [Article Influence: 76.3] [Reference Citation Analysis (0)] |
| 98. | Yamakawa M, Ito D, Honda T, Kubo K, Noda M, Nakajima K, Suzuki N. Characterization of the dipeptide repeat protein in the molecular pathogenesis of c9FTD/ALS. Hum Mol Genet. 2015;24:1630-1645. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 110] [Cited by in F6Publishing: 116] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 99. | Cleary JD, Ranum LP. Repeat-associated non-ATG (RAN) translation in neurological disease. Hum Mol Genet. 2013;22:R45-R51. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 125] [Cited by in F6Publishing: 128] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 100. | Pearson CE. Repeat associated non-ATG translation initiation: one DNA, two transcripts, seven reading frames, potentially nine toxic entities! PLoS Genet. 2011;7:e1002018. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 77] [Cited by in F6Publishing: 89] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 101. | Reddy K, Zamiri B, Stanley SY, Macgregor RB, Pearson CE. The disease-associated r(GGGGCC)n repeat from the C9orf72 gene forms tract length-dependent uni- and multimolecular RNA G-quadruplex structures. J Biol Chem. 2013;288:9860-9866. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 237] [Cited by in F6Publishing: 245] [Article Influence: 22.3] [Reference Citation Analysis (0)] |
| 102. | Mirkin EV, Mirkin SM. Replication fork stalling at natural impediments. Microbiol Mol Biol Rev. 2007;71:13-35. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 384] [Cited by in F6Publishing: 385] [Article Influence: 22.6] [Reference Citation Analysis (0)] |
| 103. | Pearson CE, Nichol Edamura K, Cleary JD. Repeat instability: mechanisms of dynamic mutations. Nat Rev Genet. 2005;6:729-742. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 671] [Cited by in F6Publishing: 642] [Article Influence: 33.8] [Reference Citation Analysis (0)] |
| 104. | Dobkin CS, Nolin SL, Cohen I, Sudhalter V, Bialer MG, Ding XH, Jenkins EC, Zhong N, Brown WT. Tissue differences in fragile X mosaics: mosaicism in blood cells may differ greatly from skin. Am J Med Genet. 1996;64:296-301. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 105. | Nolin SL, Glicksman A, Houck GE, Brown WT, Dobkin CS. Mosaicism in fragile X affected males. Am J Med Genet. 1994;51:509-512. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 113] [Cited by in F6Publishing: 111] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 106. | Thornell LE, Lindstöm M, Renault V, Klein A, Mouly V, Ansved T, Butler-Browne G, Furling D. Satellite cell dysfunction contributes to the progressive muscle atrophy in myotonic dystrophy type 1. Neuropathol Appl Neurobiol. 2009;35:603-613. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 60] [Cited by in F6Publishing: 65] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 107. | van Blitterswijk M, DeJesus-Hernandez M, Niemantsverdriet E, Murray ME, Heckman MG, Diehl NN, Brown PH, Baker MC, Finch NA, Bauer PO. Association between repeat sizes and clinical and pathological characteristics in carriers of C9ORF72 repeat expansions (Xpansize-72): a cross-sectional cohort study. Lancet Neurol. 2013;12:978-988. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 199] [Cited by in F6Publishing: 201] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 108. | Godler DE, Tassone F, Loesch DZ, Taylor AK, Gehling F, Hagerman RJ, Burgess T, Ganesamoorthy D, Hennerich D, Gordon L. Methylation of novel markers of fragile X alleles is inversely correlated with FMRP expression and FMR1 activation ratio. Hum Mol Genet. 2010;19:1618-1632. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 70] [Cited by in F6Publishing: 80] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 109. | Pretto D, Yrigollen CM, Tang HT, Williamson J, Espinal G, Iwahashi CK, Durbin-Johnson B, Hagerman RJ, Hagerman PJ, Tassone F. Clinical and molecular implications of mosaicism in FMR1 full mutations. Front Genet. 2014;5:318. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 68] [Cited by in F6Publishing: 73] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 110. | Jansen G, Willems P, Coerwinkel M, Nillesen W, Smeets H, Vits L, Höweler C, Brunner H, Wieringa B. Gonosomal mosaicism in myotonic dystrophy patients: involvement of mitotic events in (CTG)n repeat variation and selection against extreme expansion in sperm. Am J Hum Genet. 1994;54:575-585. [PubMed] [Cited in This Article: ] |
| 111. | López Castel A, Cleary JD, Pearson CE. Repeat instability as the basis for human diseases and as a potential target for therapy. Nat Rev Mol Cell Biol. 2010;11:165-170. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 338] [Cited by in F6Publishing: 325] [Article Influence: 23.2] [Reference Citation Analysis (0)] |
| 112. | Axford MM, Wang YH, Nakamori M, Zannis-Hadjopoulos M, Thornton CA, Pearson CE. Detection of slipped-DNAs at the trinucleotide repeats of the myotonic dystrophy type I disease locus in patient tissues. PLoS Genet. 2013;9:e1003866. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 48] [Cited by in F6Publishing: 49] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 113. | Cleary JD, Nichol K, Wang YH, Pearson CE. Evidence of cis-acting factors in replication-mediated trinucleotide repeat instability in primate cells. Nat Genet. 2002;31:37-46. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 137] [Cited by in F6Publishing: 151] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 114. | Nichol Edamura K, Leonard MR, Pearson CE. Role of replication and CpG methylation in fragile X syndrome CGG deletions in primate cells. Am J Hum Genet. 2005;76:302-311. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 41] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 115. | Panigrahi GB, Cleary JD, Pearson CE. In vitro (CTG)*(CAG) expansions and deletions by human cell extracts. J Biol Chem. 2002;277:13926-13934. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 42] [Cited by in F6Publishing: 49] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 116. | Panigrahi GB, Lau R, Montgomery SE, Leonard MR, Pearson CE. Slipped (CTG)*(CAG) repeats can be correctly repaired, escape repair or undergo error-prone repair. Nat Struct Mol Biol. 2005;12:654-662. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 119] [Cited by in F6Publishing: 120] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 117. | Pearson CE, Tam M, Wang YH, Montgomery SE, Dar AC, Cleary JD, Nichol K. Slipped-strand DNAs formed by long (CAG)*(CTG) repeats: slipped-out repeats and slip-out junctions. Nucleic Acids Res. 2002;30:4534-4547. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 116] [Cited by in F6Publishing: 125] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 118. | Samadashwily GM, Raca G, Mirkin SM. Trinucleotide repeats affect DNA replication in vivo. Nat Genet. 1997;17:298-304. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 248] [Cited by in F6Publishing: 248] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 119. | Slean MM, Reddy K, Wu B, Nichol Edamura K, Kekis M, Nelissen FH, Aspers RL, Tessari M, Schärer OD, Wijmenga SS. Interconverting conformations of slipped-DNA junctions formed by trinucleotide repeats affect repair outcome. Biochemistry. 2013;52:773-785. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 27] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 120. | Usdin K, Woodford KJ. CGG repeats associated with DNA instability and chromosome fragility form structures that block DNA synthesis in vitro. Nucleic Acids Res. 1995;23:4202-4209. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 178] [Cited by in F6Publishing: 200] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 121. | De Temmerman N, Seneca S, Van Steirteghem A, Haentjens P, Van der Elst J, Liebaers I, Sermon KD. CTG repeat instability in a human embryonic stem cell line carrying the myotonic dystrophy type 1 mutation. Mol Hum Reprod. 2008;14:405-412. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 23] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 122. | Seriola A, Spits C, Simard JP, Hilven P, Haentjens P, Pearson CE, Sermon K. Huntington’s and myotonic dystrophy hESCs: down-regulated trinucleotide repeat instability and mismatch repair machinery expression upon differentiation. Hum Mol Genet. 2011;20:176-185. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 67] [Cited by in F6Publishing: 73] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 123. | De Temmerman N, Sermon K, Seneca S, De Rycke M, Hilven P, Lissens W, Van Steirteghem A, Liebaers I. Intergenerational instability of the expanded CTG repeat in the DMPK gene: studies in human gametes and preimplantation embryos. Am J Hum Genet. 2004;75:325-329. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 58] [Cited by in F6Publishing: 60] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 124. | Dean NL, Tan SL, Ao A. Instability in the transmission of the myotonic dystrophy CTG repeat in human oocytes and preimplantation embryos. Fertil Steril. 2006;86:98-105. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 22] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 125. | Wöhrle D, Salat U, Hameister H, Vogel W, Steinbach P. Demethylation, reactivation, and destabilization of human fragile X full-mutation alleles in mouse embryocarcinoma cells. Am J Hum Genet. 2001;69:504-515. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 69] [Cited by in F6Publishing: 72] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 126. | Du J, Campau E, Soragni E, Ku S, Puckett JW, Dervan PB, Gottesfeld JM. Role of mismatch repair enzymes in GAA·TTC triplet-repeat expansion in Friedreich ataxia induced pluripotent stem cells. J Biol Chem. 2012;287:29861-29872. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 95] [Cited by in F6Publishing: 87] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 127. | Hick A, Wattenhofer-Donzé M, Chintawar S, Tropel P, Simard JP, Vaucamps N, Gall D, Lambot L, André C, Reutenauer L. Neurons and cardiomyocytes derived from induced pluripotent stem cells as a model for mitochondrial defects in Friedreich’s ataxia. Dis Model Mech. 2013;6:608-621. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 108] [Cited by in F6Publishing: 111] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 128. | Ku S, Soragni E, Campau E, Thomas EA, Altun G, Laurent LC, Loring JF, Napierala M, Gottesfeld JM. Friedreich’s ataxia induced pluripotent stem cells model intergenerational GAA⋅TTC triplet repeat instability. Cell Stem Cell. 2010;7:631-637. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 166] [Cited by in F6Publishing: 163] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 129. | Gerhardt J, Tomishima MJ, Zaninovic N, Colak D, Yan Z, Zhan Q, Rosenwaks Z, Jaffrey SR, Schildkraut CL. The DNA replication program is altered at the FMR1 locus in fragile X embryonic stem cells. Mol Cell. 2014;53:19-31. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 77] [Cited by in F6Publishing: 79] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 130. | Hübner K, Fuhrmann G, Christenson LK, Kehler J, Reinbold R, De La Fuente R, Wood J, Strauss JF, Boiani M, Schöler HR. Derivation of oocytes from mouse embryonic stem cells. Science. 2003;300:1251-1256. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 768] [Cited by in F6Publishing: 692] [Article Influence: 33.0] [Reference Citation Analysis (0)] |
| 131. | Nayernia K, Nolte J, Michelmann HW, Lee JH, Rathsack K, Drusenheimer N, Dev A, Wulf G, Ehrmann IE, Elliott DJ. In vitro-differentiated embryonic stem cells give rise to male gametes that can generate offspring mice. Dev Cell. 2006;11:125-132. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 390] [Cited by in F6Publishing: 334] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 132. | Itskovitz-Eldor J, Schuldiner M, Karsenti D, Eden A, Yanuka O, Amit M, Soreq H, Benvenisty N. Differentiation of human embryonic stem cells into embryoid bodies compromising the three embryonic germ layers. Mol Med. 2000;6:88-95. [PubMed] [Cited in This Article: ] |
| 133. | Reubinoff BE, Pera MF, Fong CY, Trounson A, Bongso A. Embryonic stem cell lines from human blastocysts: somatic differentiation in vitro. Nat Biotechnol. 2000;18:399-404. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2078] [Cited by in F6Publishing: 1850] [Article Influence: 77.1] [Reference Citation Analysis (0)] |
| 134. | Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145-1147. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11399] [Cited by in F6Publishing: 10061] [Article Influence: 387.0] [Reference Citation Analysis (0)] |
| 135. | Pickering SJ, Minger SL, Patel M, Taylor H, Black C, Burns CJ, Ekonomou A, Braude PR. Generation of a human embryonic stem cell line encoding the cystic fibrosis mutation deltaF508, using preimplantation genetic diagnosis. Reprod Biomed Online. 2005;10:390-397. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 105] [Cited by in F6Publishing: 90] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 136. | Verlinsky Y, Strelchenko N, Kukharenko V, Rechitsky S, Verlinsky O, Galat V, Kuliev A. Human embryonic stem cell lines with genetic disorders. Reprod Biomed Online. 2005;10:105-110. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 169] [Cited by in F6Publishing: 177] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 137. | Giudice A, Trounson A. Genetic modification of human embryonic stem cells for derivation of target cells. Cell Stem Cell. 2008;2:422-433. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 87] [Cited by in F6Publishing: 90] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 138. | Murry CE, Keller G. Differentiation of embryonic stem cells to clinically relevant populations: lessons from embryonic development. Cell. 2008;132:661-680. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1300] [Cited by in F6Publishing: 1217] [Article Influence: 76.1] [Reference Citation Analysis (0)] |
| 139. | Dvash T, Benvenisty N. Human embryonic stem cells as a model for early human development. Best Pract Res Clin Obstet Gynaecol. 2004;18:929-940. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 52] [Cited by in F6Publishing: 54] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 140. | Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861-872. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14327] [Cited by in F6Publishing: 13579] [Article Influence: 848.7] [Reference Citation Analysis (0)] |
| 141. | Chamberlain SJ, Chen PF, Ng KY, Bourgois-Rocha F, Lemtiri-Chlieh F, Levine ES, Lalande M. Induced pluripotent stem cell models of the genomic imprinting disorders Angelman and Prader-Willi syndromes. Proc Natl Acad Sci USA. 2010;107:17668-17673. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 232] [Cited by in F6Publishing: 233] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 142. | Itzhaki I, Maizels L, Huber I, Zwi-Dantsis L, Caspi O, Winterstern A, Feldman O, Gepstein A, Arbel G, Hammerman H. Modelling the long QT syndrome with induced pluripotent stem cells. Nature. 2011;471:225-229. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 794] [Cited by in F6Publishing: 834] [Article Influence: 64.2] [Reference Citation Analysis (0)] |
| 143. | Kim KY, Hysolli E, Park IH. Neuronal maturation defect in induced pluripotent stem cells from patients with Rett syndrome. Proc Natl Acad Sci USA. 2011;108:14169-14174. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 155] [Cited by in F6Publishing: 170] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 144. | Lee G, Papapetrou EP, Kim H, Chambers SM, Tomishima MJ, Fasano CA, Ganat YM, Menon J, Shimizu F, Viale A. Modelling pathogenesis and treatment of familial dysautonomia using patient-specific iPSCs. Nature. 2009;461:402-406. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 657] [Cited by in F6Publishing: 631] [Article Influence: 42.1] [Reference Citation Analysis (0)] |
| 145. | Thatava T, Kudva YC, Edukulla R, Squillace K, De Lamo JG, Khan YK, Sakuma T, Ohmine S, Terzic A, Ikeda Y. Intrapatient variations in type 1 diabetes-specific iPS cell differentiation into insulin-producing cells. Mol Ther. 2013;21:228-239. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 75] [Cited by in F6Publishing: 79] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 146. | Yang J, Cai J, Zhang Y, Wang X, Li W, Xu J, Li F, Guo X, Deng K, Zhong M. Induced pluripotent stem cells can be used to model the genomic imprinting disorder Prader-Willi syndrome. J Biol Chem. 2010;285:40303-40311. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 80] [Cited by in F6Publishing: 85] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 147. | Ma H, Morey R, O’Neil RC, He Y, Daughtry B, Schultz MD, Hariharan M, Nery JR, Castanon R, Sabatini K. Abnormalities in human pluripotent cells due to reprogramming mechanisms. Nature. 2014;511:177-183. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 259] [Cited by in F6Publishing: 251] [Article Influence: 25.1] [Reference Citation Analysis (0)] |
| 148. | Mayshar Y, Ben-David U, Lavon N, Biancotti JC, Yakir B, Clark AT, Plath K, Lowry WE, Benvenisty N. Identification and classification of chromosomal aberrations in human induced pluripotent stem cells. Cell Stem Cell. 2010;7:521-531. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 583] [Cited by in F6Publishing: 554] [Article Influence: 42.6] [Reference Citation Analysis (0)] |
| 149. | Bontekoe CJ, Bakker CE, Nieuwenhuizen IM, van der Linde H, Lans H, de Lange D, Hirst MC, Oostra BA. Instability of a (CGG)98 repeat in the Fmr1 promoter. Hum Mol Genet. 2001;10:1693-1699. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 103] [Cited by in F6Publishing: 107] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 150. | Bontekoe CJ, de Graaff E, Nieuwenhuizen IM, Willemsen R, Oostra BA. FMR1 premutation allele (CGG)81 is stable in mice. Eur J Hum Genet. 1997;5:293-298. [PubMed] [Cited in This Article: ] |
| 151. | Montermini L, Andermann E, Labuda M, Richter A, Pandolfo M, Cavalcanti F, Pianese L, Iodice L, Farina G, Monticelli A. The Friedreich ataxia GAA triplet repeat: premutation and normal alleles. Hum Mol Genet. 1997;6:1261-1266. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 143] [Cited by in F6Publishing: 154] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 152. | Peier AM, Nelson DL. Instability of a premutation-sized CGG repeat in FMR1 YAC transgenic mice. Genomics. 2002;80:423-432. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 36] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 153. | Zhao XN, Usdin K. Gender and cell-type-specific effects of the transcription-coupled repair protein, ERCC6/CSB, on repeat expansion in a mouse model of the fragile X-related disorders. Hum Mutat. 2014;35:341-349. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 24] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 154. | Bird MJ, Needham K, Frazier AE, van Rooijen J, Leung J, Hough S, Denham M, Thornton ME, Parish CL, Nayagam BA. Functional characterization of Friedreich ataxia iPS-derived neuronal progenitors and their integration in the adult brain. PLoS One. 2014;9:e101718. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 23] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 155. | Liu J, Koscielska KA, Cao Z, Hulsizer S, Grace N, Mitchell G, Nacey C, Githinji J, McGee J, Garcia-Arocena D. Signaling defects in iPSC-derived fragile X premutation neurons. Hum Mol Genet. 2012;21:3795-3805. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 103] [Cited by in F6Publishing: 106] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 156. | Marteyn A, Maury Y, Gauthier MM, Lecuyer C, Vernet R, Denis JA, Pietu G, Peschanski M, Martinat C. Mutant human embryonic stem cells reveal neurite and synapse formation defects in type 1 myotonic dystrophy. Cell Stem Cell. 2011;8:434-444. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 74] [Cited by in F6Publishing: 76] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 157. | Telias M, Segal M, Ben-Yosef D. Neural differentiation of Fragile X human Embryonic Stem Cells reveals abnormal patterns of development despite successful neurogenesis. Dev Biol. 2013;374:32-45. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 86] [Cited by in F6Publishing: 76] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 158. | Gore A, Li Z, Fung HL, Young JE, Agarwal S, Antosiewicz-Bourget J, Canto I, Giorgetti A, Israel MA, Kiskinis E. Somatic coding mutations in human induced pluripotent stem cells. Nature. 2011;471:63-67. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 956] [Cited by in F6Publishing: 922] [Article Influence: 70.9] [Reference Citation Analysis (0)] |
| 159. | Hussein SM, Batada NN, Vuoristo S, Ching RW, Autio R, Närvä E, Ng S, Sourour M, Hämäläinen R, Olsson C. Copy number variation and selection during reprogramming to pluripotency. Nature. 2011;471:58-62. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 722] [Cited by in F6Publishing: 681] [Article Influence: 52.4] [Reference Citation Analysis (0)] |
| 160. | Ji J, Ng SH, Sharma V, Neculai D, Hussein S, Sam M, Trinh Q, Church GM, McPherson JD, Nagy A. Elevated coding mutation rate during the reprogramming of human somatic cells into induced pluripotent stem cells. Stem Cells. 2012;30:435-440. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 136] [Cited by in F6Publishing: 141] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 161. | Laurent LC, Ulitsky I, Slavin I, Tran H, Schork A, Morey R, Lynch C, Harness JV, Lee S, Barrero MJ. Dynamic changes in the copy number of pluripotency and cell proliferation genes in human ESCs and iPSCs during reprogramming and time in culture. Cell Stem Cell. 2011;8:106-118. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 684] [Cited by in F6Publishing: 652] [Article Influence: 50.2] [Reference Citation Analysis (0)] |
| 162. | Lister R, Pelizzola M, Kida YS, Hawkins RD, Nery JR, Hon G, Antosiewicz-Bourget J, O’Malley R, Castanon R, Klugman S. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;471:68-73. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1202] [Cited by in F6Publishing: 1114] [Article Influence: 85.7] [Reference Citation Analysis (0)] |
| 163. | Nishino K, Toyoda M, Yamazaki-Inoue M, Fukawatase Y, Chikazawa E, Sakaguchi H, Akutsu H, Umezawa A. DNA methylation dynamics in human induced pluripotent stem cells over time. PLoS Genet. 2011;7:e1002085. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 244] [Cited by in F6Publishing: 228] [Article Influence: 17.5] [Reference Citation Analysis (0)] |
| 164. | Ruiz S, Diep D, Gore A, Panopoulos AD, Montserrat N, Plongthongkum N, Kumar S, Fung HL, Giorgetti A, Bilic J. Identification of a specific reprogramming-associated epigenetic signature in human induced pluripotent stem cells. Proc Natl Acad Sci USA. 2012;109:16196-16201. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 126] [Cited by in F6Publishing: 124] [Article Influence: 10.3] [Reference Citation Analysis (0)] |