PRMT5 regulates ovarian follicle development by facilitating Wt1 translation
Abstract
Protein arginine methyltransferase 5 (Prmt5) is the major type II enzyme responsible for symmetric dimethylation of arginine. Here, we found that PRMT5 was expressed at high level in ovarian granulosa cells of growing follicles. Inactivation of Prmt5 in granulosa cells resulted in aberrant follicle development and female infertility. In Prmt5-knockout mice, follicle development was arrested with disorganized granulosa cells in which WT1 expression was dramatically reduced and the expression of steroidogenesis-related genes was significantly increased. The premature differentiated granulosa cells were detached from oocytes and follicle structure was disrupted. Mechanism studies revealed that Wt1 expression was regulated by PRMT5 at the protein level. PRMT5 facilitated IRES-dependent translation of Wt1 mRNA by methylating HnRNPA1. Moreover, the upregulation of steroidogenic genes in Prmt5-deficient granulosa cells was repressed by Wt1 overexpression. These results demonstrate that PRMT5 participates in granulosa cell lineage maintenance by inducing Wt1 expression. Our study uncovers a new role of post-translational arginine methylation in granulosa cell differentiation and follicle development.
eLife digest
Infertility in women can be caused by many factors, such as defects in the ovaries. An important part of the ovaries for fertility are internal structures called follicles, which house early forms of egg cells. A follicle grows and develops until the egg is finally released from the ovary into the fallopian tube, where the egg can then be fertilised. In the follicle, an egg is surrounded by other types of cells, such as granulosa cells. The egg and neighbouring cells must maintain healthy contacts with each other, otherwise the follicle can stop growing and developing, potentially causing infertility.
The development of a follicle depends on an array of proteins. For example, the transcription factor WT1 controls protein levels by activating other genes and their proteins and is produced in high numbers by granulosa cells at the beginning of follicle development. Although WT1 levels dip towards the later stages of follicle development, insufficient levels can lead to defects. So far, it has been unclear how levels of WT1in granulose cells are regulated.
Chen, Dong et al. studied mouse follicles to reveal more about the role of WT1 in follicle development. The researchers measured protein levels in mouse granulosa cells as the follicles developed, and discovered elevated levels of PRMT5, a protein needed for egg cells to form and survive in the follicles. Blocking granulosa cells from producing PRMT5 led to abnormal follicles and infertility in mice. Moreover, mice that had been engineered to lack PRMT5 developed abnormal follicles, where the egg and surrounding granulosa cells were not attached to each other, and the granulosa cells had low levels of WT1. Further experiments revealed that PRMT5 controlled WT1 levels by adding small molecules called methyl groups to another regulatory protein called HnRNPA1.
The addition of methyl groups to genes or their proteins is an important modification that takes place in many processes within a cell. Chen, Dong et al. reveal that this activity also plays a key role in maintaining healthy follicle development in mice, and that PRMT5 is necessary for controlling WT1. Identifying all of the intricate mechanism involved in regulating follicle development is important for finding ways to combat infertility.
Introduction
Follicles are the basic functional units in the ovaries. Each follicle consists of an oocyte, the surrounding granulosa cells, and theca cells in the mesenchyme. The interaction between oocytes and somatic cells is crucial for follicle development. Follicle maturation experiences primordial, primary, secondary, and antral follicular stages. Primordial follicles are formed shortly after birth via breakdown of oocyte syncytia. Each primordial follicle is composed of an oocyte surrounded by a single layer of flattened pregranulosa cells that remains in a dormant phase until being recruited into the primary stage under the influence of two main signaling pathways (Monniaux, 2016). Once activated, flattened granulosa cells become cuboidal, and follicles continue to grow through proliferation of granulosa cells and enlargement of oocytes. Development of high-quality oocytes is important for female reproductive health and fertility (Jagarlamudi and Rajkovic, 2012; Liu et al., 2010; Monniaux, 2016; Richards and Pangas, 2010). Although gonadotropin, follicle-stimulating hormone (FSH), and luteinizing hormone (LH) are important for the growth of antral follicles, the early stages of follicle development are driven by a local oocyte-granulosa cell dialog. Abnormalities in this process may lead to follicle growth arrest or atresia (Oktem and Urman, 2010; Richards and Pangas, 2010).
Granulosa cells are derived from progenitors of the coelomic epithelium that direct sexual differentiation at the embryonic stage and support oocyte development postnatally (Liu et al., 2010; Smith et al., 2014). Theca-interstitial cell differentiation occurs postnatally along with the formation of secondary follicles. The steroid hormone produced by theca-interstitial cells plays important roles in follicle development and maintenance of secondary sexual characteristics (Liu et al., 2010). The Wilms’ tumor (WT) suppressor gene Wt1 is a nuclear transcription factor indispensable for normal development of several tissues. In gonads, Wt1 is mainly expressed in ovarian granulosa cells and testicular Sertoli cells. During follicle development, Wt1 is expressed at high levels in granulosa cells of primordial, primary, and secondary follicles, but its expression is decreased in antral follicles, suggesting that it might be a repressor of ovarian differentiation genes in the granulosa cells (Hsu et al., 1995). Our previous studies demonstrated that Wt1 is required for the lineage specification and maintenance of Sertoli and granulosa cells (Cen et al., 2020; Chen et al., 2017). However, the underlying mechanism that regulates the expression of Wt1 in granulosa cells is unknown.
Protein arginine methyltransferase 5 (PRMT5) is a member of the PRMT family that catalyzes the transfer of methyl groups from S-adenosylmethionine to a variety of substrates and is involved in many cellular processes, such as cell growth, differentiation, and development (Di Lorenzo and Bedford, 2011; Karkhanis et al., 2011; Stopa et al., 2015). PRMT5 is the predominant type II methyltransferase that catalyzes the formation of most symmetric dimethylarginines (SDMAs) in the cells and regulates gene expression at the transcriptional and posttranscriptional levels (Karkhanis et al., 2011). PRMT5 forms a complex with its substrate-binding partner, the WD-repeat protein MEP50 (or WDR77), which greatly enhances the methyltransferase activity of PRMT5 by increasing its affinity for protein substrates (Stopa et al., 2015).
In gonad development, inactivation of Prmt5 specifically in primordial germ cells (PGCs) causes massive loss of PGCs (Kim et al., 2014; Li et al., 2015; Wang et al., 2015). PRMT5 promotes PGC survival by regulating RNA splicing (Li et al., 2015) and suppressing transposable elements at the time of global DNA demethylation (Kim et al., 2014). In this study, we found that PRMT5 is expressed at high level in ovarian granulosa cells of growing follicles and the expression level changes with follicle development, suggesting that PRMT5 in granulosa cells plays a role in follicle development. To test the function of PRMT5 in granulosa cells, we specifically inactivated Prmt5 in granulosa cells using Sf1+/cre. The Sf1+/cre mouse expresses Cre recombinase in the adrenogonadal primordium at 10 dpc, the precursors for cortical cells in the adrenals and somatic cells in the gonads (Bingham et al., 2006; Huang and Yao, 2010). We found that Prmt5flox/flox;Sf1+/cre female mice were infertile and that follicles were arrested at the secondary stage. The expression of WT1 was dramatically reduced, and the granulosa cells in secondary follicles began to express steroidogenic genes. Further studies revealed that PRMT5 regulates follicle development by facilitating Wt1 translation.
Results
Deletion of Prmt5 in granulosa cells caused aberrant ovary development and female infertility
The expression of PRMT5 in ovarian granulosa cells was examined by immunofluorescence. As shown in Figure 1—figure supplement 1, PRMT5 (red) was expressed in oocytes, but no PRMT5 signal was detected in the granulosa cells of primordial follicles (A, A’, white arrows). PRMT5 started to be expressed in granulosa cells of primary follicles (B, B’, white arrows) and was continuously expressed in granulosa cells of secondary follicles (C, C’, white arrows), and antral follicles (D, D’, white arrows), but its expression decreased significantly in the corpus luteum (E, E’, white arrows). To test the functions of PRMT5 in granulosa cell development, we specifically deleted Prmt5 in granulosa cells by crossing Prmt5flox/flox mice with Sf1+/cre transgenic mice. In Prmt5flox/flox;Sf1+/cre female mice, PRMT5 expression was completely absent from granulosa cells (Figure 1—figure supplement 2B,D, arrows), whereas the expression of PRMT5 in oocytes and interstitial cells was not affected, suggesting that Prmt5 was specifically deleted in granulosa cells.
No obvious developmental abnormalities were observed in adult Prmt5flox/flox;Sf1+/cre mice (Figure 1A). However, the female mice were infertile with atrophic ovaries (Figure 1B, Figure 1—figure supplement 3). The results of immunohistochemistry showed growing follicles at different stages in control ovaries at 2 months of age (Figure 1C). In contrast, only a small number of follicles and few corpora lutea were observed in Prmt5flox/flox;Sf1+/cre mice (Figure 1D). We further examined follicle development in Prmt5flox/flox;Sf1+/cre mice at different developmental stages. As shown in Figure 1, a large number of growing follicles at the primary and secondary stages were observed in Prmt5flox/flox;Sf1+/cre mice (F) at 2 weeks, which was comparable to the situation in control ovaries (E). Many follicles progressed to antral follicle stages in control mice at 3 weeks (G), whereas the development of follicles in Prmt5flox/flox;Sf1+/cre mice was arrested at the secondary stage. Almost no antral follicle was observed (Figure 1—figure supplement 4B) and aberrant granulosa cells were evident (Figure 1H). The defects in follicle development were more obvious at 4 and 5 weeks (Figure 1J and L, Figure 1—figure supplement 4C and D). The number of granulosa cells around oocytes was dramatically reduced, and follicle structure was disrupted.
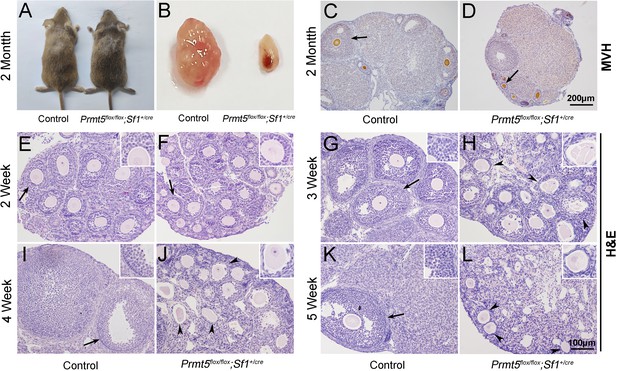
Loss of Prmt5 in granulosa cells caused aberrant follicle development and female infertility.
No developmental abnormalities were observed in Prmt5flox/flox;Sf1+/cre mice (A) at 2 months of age, and the ovary size was dramatically reduced (B). Histology of ovaries from control (C) and Prmt5flox/flox;Sf1+/cre mice (D) at 2 months of age. The histology of ovarian follicles was grossly normal in Prmt5flox/flox mice at 2 weeks (F, black arrows). Defects in follicle development were observed in Prmt5-mutant mice at 3 weeks (H, black arrowheads). Aberrant ovarian follicles with disorganized granulosa cells were observed in Prmt5flox/flox;Sf1+/cre mice at 4 (J, black arrowheads) and 5 (L, black arrowheads) weeks of age. (E), (G), (I), (K) are the histology of ovarian follicles in control mice respectively at 2, 3, 4, and 5 weeks.
The identity of granulosa cells in Prmt5flox/flox;Sf1+/cre mice was changed
To explore the underlying mechanism that caused the defects in follicle development in Prmt5flox/flox;Sf1+/cre mice, the expression of granulosa cell-specific genes was analyzed by immunohistochemistry. As shown in Figure 2, FOXL2 protein was expressed in the granulosa cells of both control (A, C) and Prmt5flox/flox;Sf1+/cre mice (B, D) at P14 and P18. WT1 protein was expressed in granulosa cells of primordial, primary, and secondary follicles in control mice at P14 and P18 (E, E’, G, G’, arrows). WT1 was also detected in the follicles of Prmt5flox/flox;Sf1+/cre mice at P14 (F, arrow). However, not all the granulosa cells were WT1-positive; some of them were WT1-negative (F’, arrowheads). The WT1 signal was almost completely absent from the majority of granulosa cells in Prmt5flox/flox;Sf1+/cre mice at P18 (H, H’); very few granulosa cells were WT1-positive (H’, arrowheads). We also found that the granulosa cells in control ovaries were cuboidal and well-organized (G’, arrow). In contrast, the granulosa cells in Prmt5flox/flox;Sf1+/cre mice were flattened (H’, dashed line circle) and were indistinguishable from surrounding stromal cells. The decreased WT1 protein expression in Prmt5-deficient granulosa cells was also confirmed by FOXL2/WT1 double staining (Figure 2—figure supplement 1).
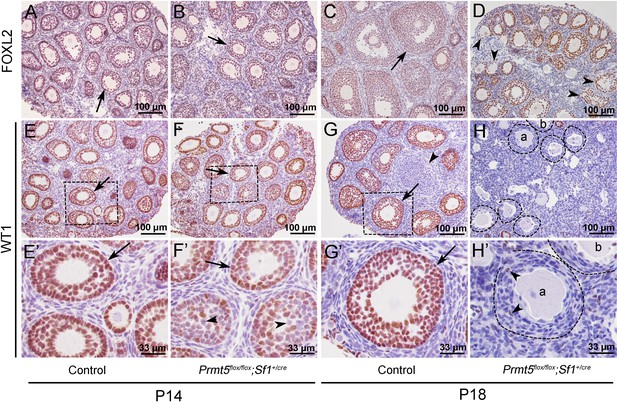
The expression of WT1 was dramatically reduced in the granulosa cells of Prmt5flox/flox;Sf1+/cre mice at P18.
The expression of FOXL2 and WT1 in granulosa cells of control and Prmt5flox/flox;Sf1+/cre mice was examined by immunohistochemistry. FOXL2 protein was expressed in the granulosa cells of both control (A, C) and Prmt5flox/flox;Sf1+/cre mice (B, D) at P14 and P18. WT1 protein was expressed in granulosa cells of primordial, primary, and secondary follicles in control mice at P14 and P18 (E, E’, G, G’, black arrows). WT1 expression was absent from most granulosa cells in Prmt5flox/flox;Sf1+/cre mice at P18 (H, H’); only very few granulosa cells were WT1-positive (H’, black arrowheads). (E’–H’) are the magnified views of (E–H) respectively.
Wt1 plays a critical role in granulosa cell development, and mutation of Wt1 leads to pregranulosa cell-to-steroidogenic cell transformation (Cen et al., 2020; Chen et al., 2017). Therefore, we further examined the expression of steroidogenic genes in Prmt5-deficient granulosa cells at P18. As shown in Figure 3, in control ovaries, 3β-HSD (3β-hydroxysteroid dehydrogenase, also known as Hsd3B1) and CYP11A1 (cytochrome P450, family 11, subfamily a, polypeptide 1, also known as P450scc) were expressed in theca-interstitial cells (A, C, arrowheads). In addition to theca-interstitial cells, 3β-HSD (B, green, arrows) and CYP11A1 (D, red, arrows) were also detected in the granulosa cells of Prmt5flox/flox;Sf1+/cre mice. We also examined the expression of SF1 (steroidogenic factor 1, also known as NR5A1), which is a key regulator of steroid hormone biosynthesis (Ikeda et al., 1993). As expected, SF1 was expressed only in theca-interstitial cells of control ovaries (E, red, arrowheads), whereas a high level of SF1 expression was detected in Prmt5-deficient granulosa cells (F, red, arrows), suggesting that the identity of granulosa cells was changed. The follicle structure was destroyed as indicated by disorganized Laminin staining (H, arrows). The proliferation and apoptosis of Prmt5-deficient granulosa cells were analyzed by BrdU incorporation and TUNEL assays. As shown in Figure 3—figure supplement 1, the numbers of TUNEL-positive cells and BrdU-positive granulosa cells were not changed in Prmt5flox/flox;Sf1+/cre ovaries compared to control ovaries at P14 and P18.
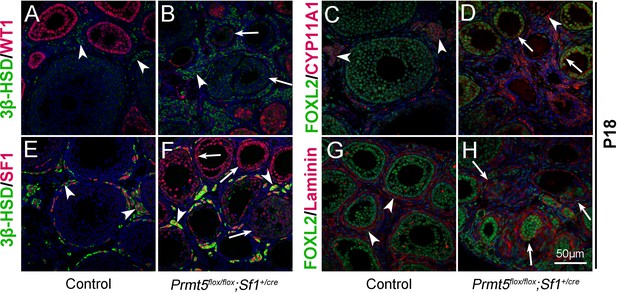
The identity of granulosa cells in Prmt5flox/flox;Sf1+/cre mice was changed.
The expression of 3β-HSD, WT1, FOXL2, CYP11A1, and SF1 in ovaries of control and Prmt5flox/flox;Sf1+/cre mice at P18 was examined by immunofluorescence. In control ovaries, 3β-HSD (A), CYP11A1 (C), and SF1 (E) were expressed only in theca-interstitial cells (white arrowheads). In the ovaries of Prmt5flox/flox;Sf1+/cre mice, 3β-HSD (B), CYP11A1 (D), and SF1 (F) were also detected in granulosa cells (white arrows). Compared to the intact follicle structure in control ovaries (arrowheads, G), the follicle structure was disordered in Prmt5flox/flox;Sf1+/cre ovaries (arrows, H) as shown by Laminin expression. DAPI (blue) was used to stain the nuclei.
To further confirm the above results, follicles were dissected from the ovaries of 2-week-old mice and cultured in vitro. As shown in Figure 3—figure supplement 2, the histology of follicles from Prmt5flox/flox;Sf1+/cre mice was comparable to that of control follicles at D2. Proliferation of granulosa cells in control follicles was observed at D4, and the follicles developed to the preovulatory stage with multiple layers of granulosa cells after 9 days of culture (A–C, G, H). The granulosa cells were detached from oocytes in Prmt5flox/flox;Sf1+/cre follicles at D4 (E, and a magnified view in L), and no colonized granulosa cells were observed after 9 days of culture (D–F, J, K). Most of the granulosa cells were attached to the culture dishes just like the interstitial cells, and denuded oocytes were observed after 3 days of culture (E, F, J, K, and a magnified view in L).
To further verify the differential expression of granulosa cell-specific and steroidogenic genes in Prmt5-deficient granulosa cells, granulosa cells were isolated at P18, and gene expression was analyzed by western blot and real-time PCR analyses. As shown in Figure 4A,B, the protein levels of PRMT5 and its interacting partner MEP50 were decreased dramatically in Prmt5flox/flox;Sf1+/cre granulosa cells, as expected. The protein level of WT1 was significantly reduced in Prmt5-deficient granulosa cells. Surprisingly, the mRNA level of Wt1 was not changed in Prmt5-deficient granulosa cells (C). FOXL2 expression was also decreased, but the difference was not significant. The expression of the steroidogenic genes CYP11A1, StAR, and NR5A1 was significantly increased in Prmt5-deficient granulosa cells, consistent with the immunostaining results. The mRNA levels of these genes were also significantly increased (C). Cyp19a1 is expressed in granulosa cells and its expression is increased in antral and preovulatory follicles (Doody et al., 1990; Stocco, 2008). We found that the mRNA level of Cyp19a1 was also significantly increased in Prmt5-deficient granulosa cells (C). We also examined the functions of PRMT5 by treating granulosa cells with the PRMT5-specific inhibitor EPZ015666. The protein level of WT1 was significantly reduced after EZP015666 treatment, whereas the mRNA level was not changed. The expression of steroidogenic genes was significantly increased at both the protein and mRNA levels after EZP015666 treatment (Figure 4D, E and F). These results were consistent with those in Prmt5-deficient granulosa cells, indicating that the effect of PRMT5 on granulosa cells was dependent on its methyltransferase activity. These results suggest that PRMT5 is required for maintenance of granulosa cell identity and that inactivation of this gene causes premature luteinization of granulosa cells.
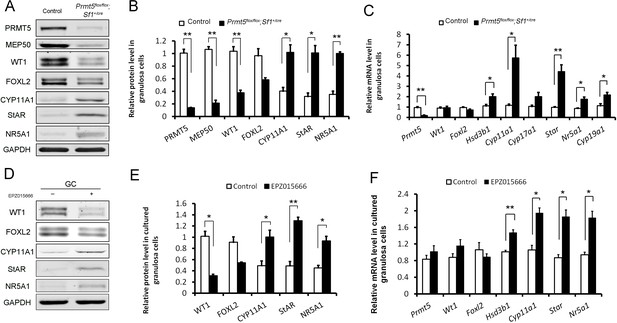
Differentially expressed genes in Prmt5-deficient granulosa cells.
Western blot (A, B) and real-time PCR analyses (C) of the indicated genes in granulosa cells isolated from control or Prmt5flox/flox;Sf1+/cre ovaries at P18. Western blot (D, E) and real-time PCR analyses (F) of the indicated genes in granulosa cells treated with DMSO or EPZ015666 (5 μM) for 5 days. The protein expression of three independent experiments in western blot analysis was quantified and normalized to that of GAPDH (B, E). The data are presented as the mean ± SEM. For (B, E, F), n = 3; for (C), n = 5. *p<0.05. **p<0.01.
-
Figure 4—source data 1
Source data for Figure 4B, C, E and F.
Differentially expressed genes in Prmt5-deficient granulosa cells.
- https://cdn.elifesciences.org/articles/68930/elife-68930-fig4-data1-v2.xlsx
The expression of WT1 was regulated by PRMT5 at the translational level
PRMT5 has been reported to regulate the translation of several genes in an IRES-dependent manner (Gao et al., 2017; Holmes et al., 2019). Internal ribosome entry sites (IRESs) are secondary structures in the 5′UTR that directly recruit the ribosome cap independently and initiate translation without cap binding and ribosome scanning (Baird et al., 2006; Coldwell et al., 2000; Stoneley and Willis, 2004). Wt1 5′UTR is 268 bp, GC-rich (68%) and contains seven CUG codons and one AUG codon. These features usually act as strong barriers for ribosome scanning and conventional translation initiation. Translation initiation in a number of these mRNAs is achieved via IRES-mediated mechanisms (Stoneley and Willis, 2004). To test whether the Wt1 5′UTR contains an IRES element, we utilized a pRF dicistronic reporter construct in which upstream Renilla luciferase is translated cap-dependently, whereas downstream Firefly luciferase is not translated unless a functional IRES is present. A stable hairpin structure upstream of Renilla luciferase minimizes cap-dependent translation (Coldwell et al., 2000, Figure 5A). Wt1 5′UTR was inserted into the intercistronic region between Renilla and Firefly luciferase (named pRWT1F), and primary granulosa cells were transfected with pRF or pRWT1F. The Firefly/Renilla luciferase activity ratio was analyzed 24 hr later. As shown in Figure 5B, the Firefly/Renilla luciferase activity ratio was dramatically increased in pRWT1F-transfected cells compared to pRF-transfected cells. In contrast, the Firefly/Renilla luciferase activity ratio was not increased when Wt1 5′UTR was inserted in the reverse direction (pRWT1-RevF; Figure 5B). The luciferase activity was dramatically increased with insertion of Ccnd1 5′UTR as a positive control, which has been reported to contain an IRES element in the 5′UTR (Shi et al., 2005). These results suggest that Wt1 5′UTR probably contains an IRES element.
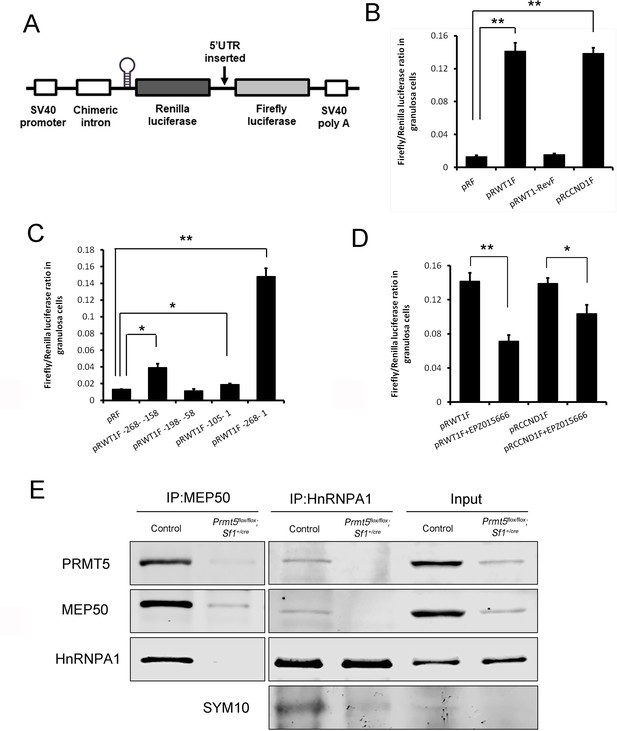
PRMT5 regulated translation of Wt1 mRNA by inducing internal ribosome entry site (IRES) activity in the 5′UTR.
(A) Schematic representation of the dicistronic reporter construct. (B) Wt1 5′UTR has IRES activity. Cultured primary granulosa cells were transfected with pRF, pRWT1F (pRF with the Wt1 5′UTR inserted), pRWT1-RevF (pRF with the Wt1 5′UTR inserted in reverse orientation), or pRCCND1F (pRF with the Ccnd1 5′UTR inserted). The Firefly and Renilla luciferase activities were measured 24 hr later, and the ratios of Firefly luciferase activity to Renilla luciferase activity were calculated. (C) The full length of Wt1 5′UTR is required for maximal luciferase activity. Three fragments of Wt1 5′UTR were inserted into pRF construct (pRWT1F –268 to –158, pRWT1F –198 –to –58, pRWT1F –105 to –1) and the constructs were transfected into primary granulosa cells. 24 hr later, the cells were harvested for luciferase activity analysis. (D) Luciferase activity was decreased in primary granulosa cells treated with the PRMT5 inhibitor EPZ015666. Isolated granulosa cells were treated with DMSO or EPZ015666 for 4 days. The day granulosa cells were isolated was denoted as day 1. On day 4, granulosa cells were transfected with pRWT1F or pRCCND1F. 24 hr later, the cells were harvested for luciferase activity analysis. The ratios of Firefly luciferase activity to Renilla luciferase activity were calculated. In (B–D), the data are presented as the mean ± SEM, n = 4. *p<0.05. **p<0.01. (E) Coimmunoprecipitation experiments were conducted in control and Prmt5flox/flox;Sf1+/cre granulosa cells. In control granulosa cells, HnRNPA1 was pulled down with an antibody against the PRMT5-associated protein MEP50; PRMT5 and MEP50 were pulled down by an HnRNPA1 antibody. The symmetric dimethylation of HnRNPA1 was significantly decreased in Prmt5flox/flox;Sf1+/cre granulosa cells. Blots are representative of three independent experiments.
-
Figure 5—source data 1
Source data for Figure 5B, C and D.
PRMT5 regulated translation of Wt1 mRNA by inducing internal ribosome entry site (IRES) activity in the 5′UTR.
- https://cdn.elifesciences.org/articles/68930/elife-68930-fig5-data1-v2.xlsx
To verify that Firefly luciferase protein was synthesized by translation of an intact dicistronic transcript instead of a monocistronic mRNA generated by cryptic splicing or promoter within the dicistronic gene (Kunze et al., 2016), mRNA from pRF- or pRWT1F-transfected cells was treated with DNase, reverse-transcribed, and then amplified with primers binding to the 5′ end of Renilla luciferase and 3′ end of Firefly luciferase open reading frame spanning the whole transcript. Only one band was detected in both cells with the expected molecular weight (Figure 5—figure supplement 1A). Moreover, qPCR analysis of Firefly and Renilla luciferase mRNA levels also showed that the Firefly/Renilla luciferase mRNA ratio was not different between pRF- and pRWT1F-transfected cells (Figure 5—figure supplement 1B), further excluding the possibility that insertion of the Wt1 5′UTR into pRF generated a monocistronic Firefly ORF.
To examine which part of Wt1 5′UTR contributes to its IRES activity, Wt1 5′UTR was divided into three fragments and respectively inserted into pRF construct (pRWT1F –268 to –158, pRWT1F –198 to –58, pRWT1F –105 to 1). These constructs were transfected into primary granulosa cells (Figure 5C). No IRES activity was detected for middle fragment (–198 to –58). Although the luciferase activity of pRWT1F –268 to –158 and pRWT1F –105 to –1 was significantly increased compared with control pRF, they were much lower than that of the full-length 5′UTR, suggesting that the full length of Wt1 5~UTR is required for maximal IRES activity.
To investigate the effect of PRMT5 on Wt1 IRES activity, granulosa cells were treated with EPZ015666 for 4 days, we found that Wt1 IRES activity was decreased significantly after EPZ015666 treatment (Figure 5D). As a positive control, the IRES activity of Ccnd1 5′UTR was also significantly decreased after EPZ015666 treatment, which was consistent with previous study (Gao et al., 2017, Figure 5D). We also checked Wt1 IRES activity in Prmt5flox/flox;Sf1+/cre granulosa cells. As expected, Wt1 IRES activity was significantly decreased in Prmt5-deficient granulosa cells compared with control granulosa cells (Figure 5—figure supplement 1C). These results indicate that PRMT5 regulates Wt1 expression at the translational level by inducing its IRES activity in granulosa cells.
Wt1 IRES activity was regulated by PRMT5 through methylation of HnRNPA1
IRES-mediated translation depends on IRES trans-acting factors (ITAFs), which function by associating with the IRES and either facilitate the assembly of initiation complexes or alter the structure of the IRES (Jo et al., 2008; Kunze et al., 2016). Heterogeneous nuclear ribonucleoprotein A1 (HnRNPA1) is a well-studied RNA binding protein that plays important roles in pre-mRNA and mRNA metabolism (Dreyfuss et al., 2002). HnRNPA1 is also an ITAF that has been reported to regulate the IRES-dependent translation of many genes, such as Ccnd1, Apaf1 (Cammas et al., 2007), Myc (Jo et al., 2008), Fgf2 (Bonnal et al., 2005), and Xiap (Lewis et al., 2007; Wall and Lewis, 2017). HnRNPA1 can be methylated by PRMT1 (Wall and Lewis, 2017) or PRMT5 (Gao et al., 2017; Holmes et al., 2019), which regulates the ITAF activity of HnRNPA1. To test whether PRMT5 interacts with HnRNPA1 in granulosa cells, coimmunoprecipitation experiments were conducted. We found that in control granulosa cells HnRNPA1 and PRMT5 were pulled down by antibody against the PRMT5 main binding partner MEP50; conversely, PRMT5 and MEP50 could be pulled down by the HnRNPA1 antibody (Figure 5E). Although HnRNPA1 protein expression was not changed between control and Prmt5flox/flox;Sf1+/cre granulosa cells, the level of symmetric dimethylation of HnRNPA1 in Prmt5flox/flox;Sf1+/cre granulosa cells was significantly reduced compared with that in control granulosa cells (Figure 5E).
To test whether HnRNPA1 functions during PRMT5-mediated Wt1 translation, HnRNPA1 was knocked down in granulosa cells via siRNA transfection. Western blot analysis results showed that HnRNPA1 protein levels were significantly decreased with siRNA transfection (Figure 6A, Figure 6—figure supplement 1). We found that WT1 protein level was increased significantly in granulosa cells after knockdown of HnRNPA1. The decreased WT1 expression in EPZ015666-treated granulosa cells was partially reversed by knockdown of HnRNPA1 (Figure 6A, Figure 6—figure supplement 1). The luciferase activity of pRWT1F was increased in granulosa cells with HnRNPA1 siRNA treatment and decreased in those with EPZ015666 treatment. The decreased luciferase activity in EPZ015666-treated granulosa cells was partially reversed by knockdown of HnRNPA1 (Figure 6B). To further confirm the effect of HnRNPA1 on Wt1 IRES activity, HnRNPA1 was overexpressed in granulosa cells, and we found that Wt1 IRES activity was significantly decreased (Figure 6D). These results indicated that as an ITAF the effect of HnRNPA1 on Wt1 IRES activity was repressive.
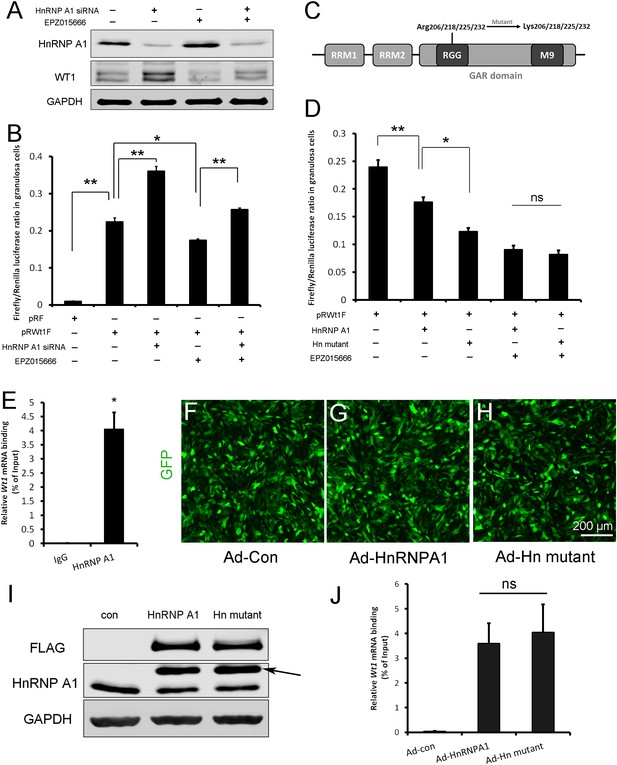
Wt1 internal ribosome entry site (IRES) activity is regulated by PRMT5 via methylation of HnRNPA1.
(A) Western blot analysis of HnRNPA1 and WT1 in granulosa cells after HnRNPA1 siRNA transfection or EPZ015666 treatment. (B) Luciferase activity analysis of pRWT1F in granulosa cells after HnRNPA1 siRNA transfection or EPZ015666 treatment. Isolated granulosa cells were treated with DMSO or EPZ015666 for 4 days. The day granulosa cells isolated is denoted as day 1. On day 2, cells were transfected with control siRNA or siRNA to HnRNPA1. 48 hr later, pRF or pRWT1F were transfected. The luciferase activity of pRWT1F was calculated as the ratio of Firefly luciferase activity to Renilla luciferase activity. (C) Schematic diagram of HnRNPA1 protein domains. HnRNPA1 contains two RNA recognition motifs (RRMs). The glycine/arginine-rich (GAR) domain contains an RGG (Arg-Gly-Gly) box and a nuclear targeting sequence (M9). Four arginine residues within the RGG motif were mutated to lysine. (D) Luciferase activity analysis of pRWT1F in granulosa cells after EPZ015666 treatment or overexpressing HnRNPA1 or arginine-mutated HnRNPA1. Isolated granulosa cells were treated with DMSO or EPZ015666 for 4 days. On day 3, flag-tagged HnRNPA1 or mutant plasmids were cotransfected with pRWT1F into granulosa cells. 48 hr later, cells were harvested for luciferase activity analysis. (E) RNA immunoprecipitation was conducted in granulosa cells using an HnRNPA1 antibody, and the Wt1 mRNA pulled down by HnRNPA1 was analyzed with real-time PCR. (F–H) Primary granulosa cells were cultured and infected with control, flag-tagged HnRNPA1, or mutant HnRNPA1 (Ad-Hn mutant) adenoviruses. The expression of control and mutant HnRNPA1 was examined by western blot analysis (I). (J) RNA immunoprecipitation was conducted using a FLAG antibody, and Wt1 mRNA pulled down by control or mutant HnRNPA1 protein was analyzed with real-time PCR. For (B, D) (n = 4) and (E, J) (n = 3), the data are presented as the mean ± SEM. *p<0.05. **p<0.01.
-
Figure 6—source data 1
Source data for Figure 6B, D, E and J.
Wt1 internal ribosome entry site (IRES) activity is regulated by PRMT5 via methylation of HnRNPA1.
- https://cdn.elifesciences.org/articles/68930/elife-68930-fig6-data1-v2.xlsx
There are five arginine residues in the HnRNPA1 glycine/arginine-rich (GAR) motif, which can be symmetrically or asymmetrically dimethylated by PRMT5 (Gao et al., 2017) or PRMT1 (Rajpurohit et al., 1994; Wall and Lewis, 2017), respectively. R206, R218, R225, and R232 are required for HnRNPA1 ITAF activity (Gao et al., 2017; Wall and Lewis, 2017). To determine the role of HnRNPA1 arginine methylation in Wt1 IRES activity, the four arginine residues were mutated to lysines (Figure 6C), and flag-tagged HnRNPA1 or mutant plasmids were cotransfected with pRWT1F into granulosa cells. We found that Wt1 IRES activity was further decreased in granulosa cells overexpressing mutant HnRNPA1 compared to those overexpressing wild-type HnRNPA1 (Figure 6D). However, the difference in Wt1 IRES activity between cells overexpressing mutant HnRNPA1 and cells overexpressing wild-type HnRNPA1 disappeared when the granulosa cells were treated with EPZ015666 (Figure 6D). These results indicate that the repressive function of HnRNPA1 on Wt1 IRES activity is inhibited by PRMT5-mediated arginine symmetric dimethylation.
To test the interaction between HnRNPA1 and Wt1 mRNA, RNA immunoprecipitation was performed with HnRNPA1 antibody in primary granulosa cells. As shown in Figure 6E, Wt1 mRNA was pulled down by the HnRNPA1 antibody in granulosa cells. Next, granulosa cells were infected with flag-tagged wild-type or arginine-mutant HnRNPA1 adenovirus (Figure 6F–I) and RNA immunoprecipitation was conducted with a FLAG antibody. The results showed that mutation of arginines did not affect the interaction between HnRNPA1 and Wt1 mRNA (Figure 6J).
The upregulation of steroidogenic genes in Prmt5flox/flox;Sf1+/cre granulosa cells was repressed by Wt1 overexpression
To test whether the upregulation of steroidogenic genes in Prmt5-deficient granulosa cells is due to downregulation of WT1, granulosa cells from Prmt5flox/flox;Sf1+/cre mice were infected with control or GFP-tagged WT1-expressing adenovirus (Figure 7A and B). Wt1 protein (Figure 7D arrow, E) and mRNA (Figure 7C) levels were dramatically increased in Prmt5-deficient granulosa cells after Wt1 overexpression. We found that the expression of steroidogenic genes was significantly decreased in these cells. These results suggest that the aberrant differentiation of Prmt5-deficient granulosa cells can be rescued by WT1.
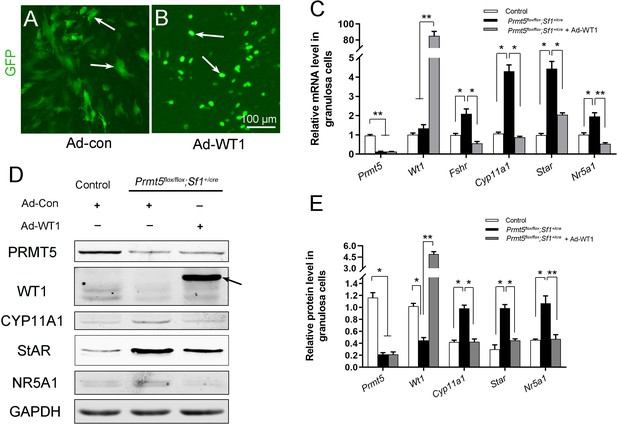
The upregulation of steroidogenic genes in Prmt5flox/flox;Sf1+/cre granulosa cells was reversed by Wt1 overexpression.
(A, B) Granulosa cells isolated from control and Prmt5flox/flox;Sf1+/cre mice were cultured and infected with control or GFP-fused Wt1 adenovirus. The expression of steroidogenic genes was examined by RT-qPCR (C) and western blot analysis (D). The protein expression of three independent experiments in western blot analysis was quantified and normalized to that of GAPDH (E). (C, E) The data are presented as the mean ± SEM (n = 3). *p<0.05. **p<0.01.
-
Figure 7—source data 1
Source data for Figure 7C and E.
The upregulation of steroidogenic genes in Prmt5flox/flox;Sf1+/cre granulosa cells was reversed by Wt1 overexpression.
- https://cdn.elifesciences.org/articles/68930/elife-68930-fig7-data1-v2.xlsx
Discussion
Protein arginine methylation is one of the most important epigenetic modifications and is involved in many cellular processes. In this study, we found that protein arginine methylation plays important roles in granulosa cell development. The development of ovarian follicles is a dynamic process. The granulosa cells in antral follicles express gonadotropin receptors. Before ovulation, granulosa cells begin to express steroidogenic enzymes that are necessary for progesterone and estradiol synthesis (Irving-Rodgers et al., 2004; Smith et al., 2014). In this study, we found that Prmt5-deficient granulosa cells began to express steroidogenic genes in secondary follicles and the upregulation of the steroidogenic genes in Prmt5-deficient granulosa cells was reversed by Wt1 overexpression, indicating that PRMT5 is required for preventing the premature differentiation of granulosa cells via regulation of WT1 expression. Coordinated interaction between granulosa cells and oocytes is required for successful follicle development and production of fertilizable oocytes. The premature luteinized granulosa cells will lose their structural and nutritional support for oocytes, which will lead to follicle growth arrest or atresia at early stages of folliculogenesis.
Nuclear receptor Sf1 plays a critical role in the regulation of steroid hormone biosynthesis by inducing the expression of steroidogenic enzymes in steroidogenic cells (Ikeda et al., 1993). Our previous study demonstrated that WT1 represses Sf1 expression by directly binding to the Sf1 promoter region and that inactivation of Wt1 causes upregulation of Sf1, which in turn activates the steroidogenic program (Chen et al., 2017). In the present study, the mRNA and protein levels of Sf1 were significantly upregulated after WT1 loss. Therefore, we speculate that the upregulation of steroidogenic genes in Prmt5-deficient granulosa cells is most likely due to the increased expression of Sf1 gene.
As an important nuclear transcription factor, the function of WT1 in granulosa cell development has been investigated. However, the molecular mechanism that regulates the expression of Wt1 gene is unknown. In this study, we found that the expression of WT1 at the protein level was dramatically reduced in Prmt5-deficient granulosa cells, whereas the mRNA level was not changed, indicating that PRMT5 regulates Wt1 expression at the post-transcriptional level. In our mouse model, Prmt5 was inactivated in granulosa cells at the early embryonic stage. However, defects in follicle development were not observed until 2 weeks after birth. This outcome probably occurred because Prmt5 is not expressed in granulosa cells before the development of primary follicles (Figure 1—figure supplement 1). During the early stage, Wt1 expression is also maintained in pregranulosa cells. Therefore, we speculate that there must be other factor(s) involved in regulating Wt1 expression before primary follicle stage.
More than 100 mRNAs in mammals contain IRES elements in their 5′UTRs (Jaud et al., 2019), which are involved in various physiological processes, such as differentiation, cell cycle progression, apoptosis, and stress responses (Godet et al., 2019). The 5′UTR sequence of Wt1 mRNA is highly conserved, with more than 85% homology among the sequences of 29 mammalian species. Our study indicates that the Wt1 5′UTR has IRES activity. HnRNPA1 belongs to the HnRNP family, which comprises at least 20 members associated with RNA processing, splicing, transport, and metabolism (Godet et al., 2019; Roy et al., 2017). As a main ITAF, HnRNPA1 either activates the translation of Fgf2 (Bonnal et al., 2005), Srebp-1a (Damiano et al., 2013), and Ccnd1 (Shi et al., 2005) or inhibits the translation of Xiap (Lewis et al., 2007), Apaf (Cammas et al., 2007), and Bcl-xl (Bevilacqua et al., 2010). The underlying mechanism by which HnRNPA1 activates some IRESs but suppresses other IRESs is still unknown. HnRNPA1 may compete with other ITAFs for binding or may modify IRES structure and thus regulate IRES activity (Cammas et al., 2007; Lewis et al., 2007).
It has been reported that the expression of several genes is regulated by PRMT5 at the protein level (Gao et al., 2017; Nicholas et al., 2013). Gao et al. reported that PRMT5 regulates IRES-dependent translation via methylation of HnRNPA1 in the 293T and MCF-7 cell lines. They found that HnRNPA1 activates the IRES-dependent translation and that methylation of HnRNPA1 facilitates the interaction of HnRNPA1 with IRES mRNA to promote translation (Gao et al., 2017). In the present study, we found that Wt1 IRES activity was repressed by HnRNPA1 (Figure 8A) and that the repressive effect of HnRNPA1 was reversed by PRMT5-mediated arginine methylation; thus, Wt1 IRES-dependent translation was promoted by PRMT5 (Figure 8B).
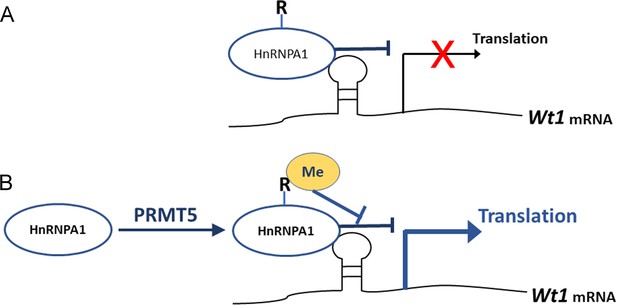
Schematic illustration of how PRMT5 regulates Wt1 mRNA translation.
(A) As an ITAF, HnRNPA1 binds to Wt1 mRNA and inhibits the internal ribosome entry site (IRES)-dependent translation of Wt1. (B) PRMT5 catalyzes symmetric methylation of HnRNPA1, which suppresses the ITAF activity of HnRNPA1 and promotes the translation of Wt1 mRNA. R: arginine. Me: methylation.
The ITAF activity of HnRNPA1 can be regulated by post-translational modifications (Godet et al., 2019). Phosphorylation of HnRNPA1 on serine 199 by Akt inhibits IRES-dependent translation of c-myc and cyclin D1 (Jo et al., 2008; Shi et al., 2005). Symmetric dimethylation of HnRNPA1 by PRMT5 enhances HnRNPA1 ITAF activity and promotes the translation of target mRNAs (Gao et al., 2017). Asymmetric dimethylation of HnRNPA1 by PRMT1 inhibits its ITAF activity (Wall and Lewis, 2017). These results suggest that arginine methylation has different effects on the ITAF activity of HnRNPA1 according to different IRESs and cell contexts. Our study demonstrated that HnRNPA1 ITAF activity toward Wt1 mRNA was repressed by PRMT5-mediated arginine methylation. However, the affinity between HnRNPA1 and Wt1 mRNA was not affected after mutation of arginine residues, consistent with the findings of a previous study (Wall and Lewis, 2017). Therefore, the inhibition of HnRNPA1 ITAF activity by PRMT5 does not occur through changes in the binding of HnRNPA1 to Wt1 mRNA. Arginine–glycine–glycine (RGG)-motif region is also reported to be involved in mediating the interactions between homo- and heterotypic proteins. It is possible that arginine methylation of HnRNPA1 changes the interactions of HnRNPA1 and its protein partners, which affects the ITAF activity of HnRNPA1 (Wall and Lewis, 2017). The underlying mechanism needs further investigation.
Epigenetic modification is involved in numerous cellular processes. However, the functions of epigenetic modification in granulosa cell development have not been well studied. In this study, we demonstrated that Prmt5 is required for maintenance of granulosa cell identity in follicle development and that inactivation of Prmt5 causes premature luteinization of granulosa cells. Our study also demonstrates that PRMT5 regulates WT1 expression at the translational level by methylating HnRNPA1. This study provides very important information for better understanding the regulation of gonad somatic cell differentiation.
Materials and methods
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (Mus musculus) | Sf1+/cre | Gift from Prof. Humphrey Hung-Chang Yao | Parker lab | Bingham et al., 2006 |
| Recombinant DNA reagent | pRF (plasmid) | Gift from Prof. Anne E Willis | Willis lab | Coldwell et al., 2000 |
| Antibody | Anti-PRMT5 (rabbit polyclonal) | Millipore | Cat# 07-405 | IF (1:200),WB (1:1000) |
| Antibody | Anti-MEP50 (rabbit monoclonal) | Abcam | Cat# ab154190 | WB (1:1000),IP (1 μg/mg protein) |
| Antibody | Anti-WT1 (rabbit monoclonal) | Abcam | Cat# ab89901 | IHC (1:400), IF (1:200), WB (1:1000) |
| Antibody | Anti-FOXL2 (goat polyclonal) | Abcam | Cat# Abcam, ab5096 | IF (1:100), WB (1:800) |
| Antibody | Anti-HnRNPA1 (mouse monoclonal) | Abcam | Cat# ab5832 | WB (1:1000)IP (1 μg/mg protein) |
| Antibody | Anti-SYM10(rabbit polyclonal) | Millipore | Cat# 07-412 | WB (1:800) |
| Antibody | Anti-CYP11A1 (rabbit polyclonal) | Proteintech | Cat# 13363-1-AP | IF (1:100), WB (1:800) |
| Antibody | Anti-SF1 (rabbit polyclonal) | Proteintech | Cat# 18658-1-AP | IF (1:200), WB (1:800) |
| Antibody | Anti-StAR (rabbit polyclonal) | Santa Cruz | Cat# sc-25806 | IF (1:100), WB (1:800) |
| Antibody | Anti-FLAG (mouse monoclonal) | Sigma-Aldrich | Cat# F1804 | WB (1:1000)RIP (1 μg/mg protein) |
| Antibody | Anti-IgG | Santa Cruz | Cat# sc-2025 | RIP (1 μg/mg protein) |
| Antibody | Anti-3β-HSD (goat polyclonal) | Santa Cruz | Cat# sc-30820 | IF (1:200), WB (1:1000) |
| Antibody | FITC-conjugated donkey anti-goat IgG | Jackson | Cat# 705-095-147 | 1:150 |
| Antibody | Cy3-conjugated donkey anti-rabbit IgG | Jackson | Cat# 711-165-152 | 1:300 |
| Commercial assay or kit | Collagenase IV | VETEC | Cat# V900893 | 1 mg/ml |
| Commercial assay or kit | Hyaluronidase | Sigma Aldrich | Cat# SIAL-H3506 | 1 mg/ml |
| Commercial assay or kit | DNase I | AppliChem | Cat# A37780500 | 1 mg/ml |
| Commercial assay or kit | EPZ015666 | MedChemExpress | Cat# HY-12727 | 5 μM |
| Commercial assay or kit | RNeasy Kit | Aidlab | Cat# RN28 | |
| Commercial assay or kit | Dual luciferase reporter assay system | Promega | Cat# E1910 | |
| Commercial assay or kit | siRNA to HnRNPA1 | ThermoFisher Scientific | Cat# S67643, S67644 | |
| Commercial assay or kit | Protein A agarose beads | GE | Cat# 17-5280-01 | |
| Commercial assay or kit | Protein G agarose beads | GE | Cat# 17-0618-01 |
Mice
All animal experiments were carried out in accordance with the protocols approved by the Institutional Animal Care and Use Committee (IACUC) of the Institute of Zoology, Chinese Academy of Sciences (CAS; SYXK 2018-0021). All mice were maintained on a C57BL/6;129/SvEv mixed background. Prmt5flox/flox;Sf1+/cre female mice were obtained by crossing Prmt5flox/flox mice with Prmt5+/flox;Sf1+/cre mice. Prmt5flox/flox and Prmt5+/flox female mice were used as controls.
For breeding experiment, both control and Prmt5flox/flox;Sf1+/cre female mice were crossed with wild-type male mice when they reached 8 weeks of age. Each pair was kept in a cage for 4 months. The number of pups delivered during this period was counted.
Plasmid and adenovirus
Request a detailed protocolThe dicistronic construct pRF was a generous gift from Professor Anne Willis, University of Cambridge. pRWT1F, pRCCND1F, and pRWT1-RevF were constructed by inserting the mouse Wt1 5′UTR, human Ccnd1 5′UTR, or mouse Wt1 5′UTR in reverse orientation into EcoRI and NcoI sites of the pRF vector. Mouse Wt1 5′UTR and human Ccnd1 5′UTR sequence were amplified by PCR. The primers amplifying the whole transcript of pRF binding to the 5′ end of Renilla and 3′ end of Firefly ORF: pRF-F: GCCACCATGACTTCGAAAGTTTATGA; pRF-R: TTACACGGCGATCTTTCCGC. FLAG-tagged HnRNPA1 and mutant plasmids were generated by inserting the coding sequence and a mutant sequence of mouse HnRNPA1, respectively, into NheI and BamHI sites of the pDC316-mCMV-ZsGreen-C-FLAG vector. Adenoviruses containing WT1 coding sequence, HnRNPA1, or the mutant sequence were generated using the Gateway Expression System (Invitrogen). The primers used for constructing the plasmids are as follows:
| Plasmid symbol | Primer 5′ to 3′ |
|---|---|
| pRWT1F-forward pRWT1F-reverse | CCGGAATTCTGTGTGAATGGAGCGGCCGAGCATCTAGCCATGGGATCGCGGCGAGGAGGCG |
| pRWT1-RevF-forward | CTAGCCATGGTGTGTGAATGGAGCGGCCGAGCAT |
| pRWT1-RevF-reverse | CCGGAATTCGATCGCGGCGAGGAGGCG |
| pRCCND1F-forward | GCTGAATTCCACACGGACTACAGGGGAGTTTT |
| pRCCND1F-reverse pRWT1F –268 to –158-forward | CGGCCATGGGGCTGGGGCTCTTCCTGGGCCCGGAATTCTGTGTGAATGGAGCGGCCGAG |
| pRWT1F –268 to –158-reverse | CTAGCCATGGACTCCTTACCCCAGCTGCCT |
| pRWT1F –198 to –58-forward | CCGGAATTCTTTGGGAAGCTGGGGGCAG |
| pRWT1F –198 to –58-reverse pRWT1F –105 to 1-forward pRWT1F –105 to 1-reverseHnRNPA1-forwardHnRNPA1-reverse | CTAGCCATGGGGTGGGTGAATGAGTGGGTCCGGAATTCTCCCCCTTTCCTTTTCCCGCCCCTAGCCATGGGATCGCGGCGAGGAGGCGCTAGCTAGCCACCATGTCTAAGTCCGAGTCTCCCAAGGACGCGGATCCGAACCTCCTGCCACTGCCATAGCTA |
Isolation of granulosa cells, transient transfection, infection, and luciferase assay
Request a detailed protocolGranulosa cells were isolated from mice at 16–18 days old. After mechanical dissection, ovaries were cut into several parts and incubated in PBS containing 1 mg/ml collagenase IV (VETEC, V900893) in a water bath with circular agitation (85 rpm) for 5 min at 37°C. Follicles were allowed to settle and washed in PBS. The supernatant were discarded. A second enzyme digestion was performed in PBS containing 1 mg/ml collagenase IV, 1 mg/ml hyaluronidase (SIGMA, SIAL-H3506), 0.25% Trypsin, and 1 mg/ml DNase I (AppliChem, A37780500) for 15 min. FBS was added to stop the digestion and cell suspension was filtered through a 40 μm filter. Cells were centrifuged, washed, and then plated in 24-well plate in DMEM/F12 supplemented with 5% FBS. For EPZ015666 treatment, granulosa cells were incubated in the medium with the addition of 5 μM EPZ015666 (MedChemExpress, HY-12727) for 4–5 days. When cells were approximately 70% confluent, granulosa cells were transfected with plasmids or infected with adenovirus according to the experiments. At the end of culture, cells were lysed for RT-qPCR, western blot analysis, or luciferase activity analysis using a dual luciferase reporter assay system (Promega, E1910).
Control siRNA or siRNA to HnRNPA1 was purchased from ThermoFisher (S67643, S67644) and transfected into granulosa cells with Lipofectamine 3000 transfection reagent without P3000. 48 hr later, pRF or pRWT1F were transfected and luciferase activities were measured the following day.
In vitro ovarian follicle culture
Request a detailed protocolFollicles were dissected and cultured as previously described (Gao et al., 2014). Briefly, ovaries of 14-day-old mice were dissected aseptically using the beveled edges of two syringe needles. Follicles with 2–3 layers of granulosa cells, a centrally placed oocyte, an intact basal membrane, and attached theca cells were selected and cultured individually in 20 μl droplets of culture medium (αMEM supplemented with 5% FBS, 1% ITS, and 100 mIU/ml recombinant FSH). The culture were maintained in 37°C and 5% CO2 in air. The medium was replaced every other day. The histology of the follicles was recorded daily under a microscope.
Coimmunoprecipitation
Request a detailed protocolGranulosa cells isolated from mice at 16–18 days old were cultured in 10 cm dishes and lysed with lysis buffer (50 mM Tris–HCl [pH 7.5], 150 mM NaCl, 1 mM EDTA, 1% Nonidet P-40) supplemented with protease inhibitors cocktail (Roche) and 1 mM PMSF. 1 mg of protein were first pre-cleared with protein A/G agarose beads (GE, 17-0618-01, 17-5280-01) for 1 hr at 4°C, then incubated with HnRNPA1 antibody (Abcam, ab5832), or MEP50 antibody (Abcam, ab154190) for 4 hr at 4°C. Then protein A and G agarose beads were added and incubated overnight. The immunoprecipitates were washed four times in lysis buffer supplemented with cocktail and PMSF, resolved in loading buffer, incubated for 5 min at 95°C, and then analyzed by western blotting. The antibodies used in western blotting include PRMT5 (Millipore, 07-405), SYM10 (Millipore, 07-412), HnRNPA1 (Abcam, ab5832), and MEP50 (Abcam, ab154190).
Western blot analysis
Request a detailed protocolGranulosa cells were washed with PBS, lysed with RIPA buffer (50 mM Tris–HCl [pH 7.5], 150 mM NaCl, 1% NP-40, 0.1% SDS, 1% sodium deoxycholate, 5 mM EDTA) supplemented with protease inhibitors cocktail (Roche) and 1 mM PMSF. Equal amounts of total protein were separated by SDS/PAGE gels, transferred to nitrocellulose membrane, and probed with the primary antibodies. The images were captured with the ODYSSEY Sa Infrared Imaging System (LI-COR Biosciences, Lincoln, NE). Densitometry was performed using ImageJ software. The protein expression was normalized to that of GAPDH. Blots are representative of three independent experiments. The antibodies used were PRMT5 (Millipore, 07-405), MEP50 (Abcam, ab154190), WT1 (Abcam, ab89901), FOXL2 (Abcam, ab5096), CYP11A1 (Proteintech, 13363-1-AP), StAR (Santa Cruz, sc-25806), SF1 (Proteintech, 18658-1-AP), and FLAG (Sigma, F1804).
RNA immunoprecipitation
Request a detailed protocolGranulosa cells were isolated from mice at 16–18 days old and cultured in 10 cm dishes. The cells were then lysed with RIP buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 5 mM EDTA, 1% NP-40, 0.5% sodium deoxycholate) supplemented with protease inhibitor cocktail and 200 U/ml RNase inhibitor. 5% of the cell lysate supernatants were used as the input and the remaining were incubated with 1.5 μg of IgG (mouse, Santa Cruz, sc-2025), HnRNPA1 antibody (Abcam, ab5832), or FLAG antibody (Sigma, F1804) for 4 hr at 4°C. Then protein A and G agarose beads were added to immunoprecipitate the RNA/protein complex. The conjugated beads were thoroughly washed with lysis buffer (50 mM Tris–HCl [pH 7.5], 500 mM NaCl, 5 mM EDTA, 1% NP-40, 0.5% sodium deoxycholate) supplemented with cocktail and 200 U/ml RNase inhibitor. Bound RNA was extracted using a RNeasy Kit and analyzed with RT-qPCR analysis.
Real-time RT-PCR
Request a detailed protocolTotal RNA was extracted using a RNeasy Kit (Aidlab, RN28) in accordance with the manufacturer’s instructions. 1 µg of total RNA was used to synthesize first-strand cDNA (Abm, G592). cDNAs were diluted and used for the template for real-time SYBR Green assay. Gapdh was used as an endogenous control. All gene expression was quantified relative to Gapdh expression. The relative concentration of the candidate gene expression was calculated using the formula 2-ΔΔCT. Real-time RT-PCR primers are as follows:
| Gene symbol | RT forward primer 5′ to 3′ | RT reverse primer 5′ to 3′ |
|---|---|---|
| Prmt5Wt1 | TGGTGGCATAACTTTCGGACTCAAGGACTGCGAGAGAAGGTTT | TCCAAGCCAGCGGTCAATTGGTGTGGGTCTTCAGATGGT |
| Hsd3b1 | CTCAGTTCTTAGGCTTCAGCAATTAC | CCAAAGGCAAGATATGATTTAGGA |
| Cyp11a1 | CCAGTGTCCCCATGCTCAAC | TGCATGGTCCTTCCAGGTCT |
| Cyp17a1 | GCCCAAGTCAAAGACACCTAAT | GTACCCAGGCGAAGAGAATAGA |
| StARSf1 | CCGGAGCAGAGTGGTGTCACCCAAGAGTTAGTGCTCCAGT | CAGTGGATGAAGCACCATGCCTGGGCGTCCTTTACGAGG |
| Foxl2Fshr | ACAACACCGGAGAAACCAGACATGTGTTCTCCAACCTACCCA | CGTAGAACGGGAACTTGGCTAGCTGGCAAGTGTTTAATGCCTG |
| Gapdh | GTCATTGAGAGCAATGCCAG | GTGTTGCTACCCCCAATGTG |
| RenillaFireflyCyp19a1 | CGTGGAAACCATGTTGCCATCAAGGTTCCATCTGCCAGGTATCAGGAACCCCATGCAGTATAATGTCAC | ACGGGATTTCACGAGGCCATGATACGTCTTCGTCCCAGTAAGCTATGAGGACCTGGTATTGAAGACGAG |
Immunohistochemistry and immunofluorescence analysis
Request a detailed protocolImmunohistochemistry procedures were performed as described previously (Gao et al., 2006). Stained sections were examined with a Nikon microscope, and images were captured by a Nikon DS-Ri1 CCD camera. For immunofluorescence analysis, the 5 μm sections were incubated with 5% BSA in 0.3% Triton X-100 for 1 hr after rehydration and antigen retrieval. The sections were then incubated with the primary antibodies for 1.5 hr and the corresponding FITC-conjugated donkey anti-goat IgG (1:150, Jackson ImmunoResearch, 705-095-147) and Cy3-conjugated donkey anti-rabbit IgG (1:300, Jackson ImmunoResearch, 711-165-152) for 1 hr at room temperature. The following primary antibodies were used: WT1 (Abcam, ab89901), FOXL2 (Abcam, ab5096), CYP11A1 (Proteintech, 13363-1-AP), SF1 (Proteintech, 18658-1-AP), and 3β-HSD (Santa Cruz, sc-30820). After being washed three times in PBS, the nuclei were stained with DAPI. The sections were examined with a confocal laser scanning microscope (Carl Zeiss Inc, Thornwood, NY).
For follicle counting analysis, whole ovaries from control and Prmt5flox/flox;Sf1+/cre female mice at 2, 3, 4, and 5 weeks of age were serially sectioned at 5 μm thickness (n = 3/time point/genotype), and follicles were counted on every five sections.
Statistical analysis
Request a detailed protocolAll experiments were repeated at least three times. 3–5 mice for each genotype at each time point were used for immunostaining or quantitative experiments. For immunostaining, one representative picture of similar results from 3 to 5 mice for each genotype at each time point is presented. The quantitative results are presented as the mean ± SEM. All granulosa cell culture experiments were repeated at least three times by using three different cell preparations. Statistical analyses were conducted using GraphPad Prism version 9.0.0. Unpaired two-tailed Student’s t-tests were used for comparison between two groups. For three or more groups, data were analyzed using one-way ANOVA. p-Values<0.05 were considered to indicate significance.
Data availability
Our work did not generate any datasets or use any previously published datasets. Source data for Figures 4, 5, 6, 7 and figure supplement of Figures 1, 5, 6 have been provided.
References
-
Eif2alpha phosphorylation tips the balance to apoptosis during osmotic stressThe Journal of Biological Chemistry 285:17098–17111.https://doi.org/10.1074/jbc.M110.109439
-
Cytoplasmic relocalization of heterogeneous nuclear ribonucleoprotein a1 controls translation initiation of specific mrnasMolecular Biology of the Cell 18:5048–5059.https://doi.org/10.1091/mbc.e07-06-0603
-
Expression of messenger ribonucleic acid species encoding steroidogenic enzymes in human follicles and corpora lutea throughout the menstrual cycleThe Journal of Clinical Endocrinology and Metabolism 70:1041–1045.https://doi.org/10.1210/jcem-70-4-1041
-
Messenger-RNA-binding proteins and the messages they carryNature Reviews. Molecular Cell Biology 3:195–205.https://doi.org/10.1038/nrm760
-
WT1 functions in ovarian follicle development by regulating granulosa cell differentiationHuman Molecular Genetics 23:333–341.https://doi.org/10.1093/hmg/ddt423
-
PRMT5 regulates ires-dependent translation via methylation of Hnrnp A1Nucleic Acids Research 45:4359–4369.https://doi.org/10.1093/nar/gkw1367
-
IRES Trans-Acting Factors, Key Actors of the Stress ResponseInternational Journal of Molecular Sciences 20:924.https://doi.org/10.3390/ijms20040924
-
Oogenesis: Transcriptional regulators and mouse modelsMolecular and Cellular Endocrinology 356:31–39.https://doi.org/10.1016/j.mce.2011.07.049
-
Heterogeneous nuclear ribonucleoprotein A1 regulates cyclin d1 and C-MYC internal ribosome entry site function through AKT signalingThe Journal of Biological Chemistry 283:23274–23287.https://doi.org/10.1074/jbc.M801185200
-
Versatility of prmt5-induced methylation in growth control and developmentTrends in Biochemical Sciences 36:633–641.https://doi.org/10.1016/j.tibs.2011.09.001
-
SST2 translation is regulated by fgf2 via an HNRNP a1-mediated ires-dependent mechanismBiochimica et Biophysica Acta 1859:848–859.https://doi.org/10.1016/j.bbagrm.2016.05.005
-
Subcellular relocalization of a trans-acting factor regulates XIAP ires-dependent translationMolecular Biology of the Cell 18:1302–1311.https://doi.org/10.1091/mbc.e06-06-0515
-
Building pathways for ovary organogenesis in the mouse embryoCurrent Topics in Developmental Biology 90:263–290.https://doi.org/10.1016/S0070-2153(10)90007-0
-
Understanding follicle growth in vivoHuman Reproduction 25:2944–2954.https://doi.org/10.1093/humrep/deq275
-
Enzymatic methylation of recombinant heterogeneous nuclear RNP protein A1. Dual substrate specificity for S-adenosylmethionine:histone-arginine N-methyltransferaseThe Journal of Biological Chemistry 269:1075–1082.
-
Handbook of Experimental Pharmacology3–27, New insights into ovarian function, Handbook of Experimental Pharmacology, Berlin/Heidelberg, Germany, Springer, 10.1007/978-3-642-02062-9_1, 20839083.
-
Emerging roles of HNRNPA1 in modulating malignant transformationWiley Interdisciplinary Reviews RNA 8:1431.https://doi.org/10.1002/wrna.1431
-
Development of mammalian ovaryThe Journal of Endocrinology 221:R145–R161.https://doi.org/10.1530/JOE-14-0062
-
The PRMT5 arginine methyltransferase: Many roles in development, cancer and beyondCellular and Molecular Life Sciences 72:2041–2059.https://doi.org/10.1007/s00018-015-1847-9
Article and author information
Author details
Funding
Ministry of Science and Technology of the People's Republic of China (2018YFC1004200)
- Min Chen
Ministry of Science and Technology of the People's Republic of China (2018YFA0107700)
- Fei Gao
Chinese Academy of Sciences (XDB19000000)
- Fei Gao
National Science Fund for Distinguished Young Scholars (81525011)
- Fei Gao
National Natural Science Foundation of China (31970785)
- Fei Gao
National Natural Science Foundation of China (31601193)
- Fei Gao
National Natural Science Foundation of China (31671496)
- Fei Gao
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Acknowledgements
We thank Prof. Anne E Willis (University of Cambridge) for her generous gift of the dicistronic contruct pRF. We thank Prof. Humphrey Hung-Chang Yao (NIEHS/NIH) for the Sf1+/cre mice. This work was supported by the National Key R&D Program of China (2018YFC1004200, 2018YFA0107700); Strategic Priority Research Program of the Chinese Academy of Sciences (XDB19000000); the National Science Fund for Distinguished Young Scholars (81525011); and the National Natural Science Foundation of China (31970785, 31601193, and 31671496).
Ethics
This study was performed in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of Institute of Zoology, Chinese Academy of Sciences (CAS). All animal experiments were carried out in accordance with the protocols approved by the Institutional Animal Care and Use Committee at the Institute of Zoology, Chinese Academy of Sciences (CAS) (Permit Number: SYXK 2018-0021).
Version history
- Received: March 30, 2021
- Preprint posted: April 7, 2021 (view preprint)
- Accepted: August 26, 2021
- Accepted Manuscript published: August 27, 2021 (version 1)
- Version of Record published: September 30, 2021 (version 2)
Copyright
© 2021, Chen et al.
This article is distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use and redistribution provided that the original author and source are credited.
Metrics
-
- 1,481
- views
-
- 298
- downloads
-
- 10
- citations
Views, downloads and citations are aggregated across all versions of this paper published by eLife.
Download links
Downloads (link to download the article as PDF)
Open citations (links to open the citations from this article in various online reference manager services)
Cite this article (links to download the citations from this article in formats compatible with various reference manager tools)
Further reading
-
- Cell Biology
- Neuroscience
Alternative RNA splicing is an essential and dynamic process in neuronal differentiation and synapse maturation, and dysregulation of this process has been associated with neurodegenerative diseases. Recent studies have revealed the importance of RNA-binding proteins in the regulation of neuronal splicing programs. However, the molecular mechanisms involved in the control of these splicing regulators are still unclear. Here, we show that KIS, a kinase upregulated in the developmental brain, imposes a genome-wide alteration in exon usage during neuronal differentiation in mice. KIS contains a protein-recognition domain common to spliceosomal components and phosphorylates PTBP2, counteracting the role of this splicing factor in exon exclusion. At the molecular level, phosphorylation of unstructured domains within PTBP2 causes its dissociation from two co-regulators, Matrin3 and hnRNPM, and hinders the RNA-binding capability of the complex. Furthermore, KIS and PTBP2 display strong and opposing functional interactions in synaptic spine emergence and maturation. Taken together, our data uncover a post-translational control of splicing regulators that link transcriptional and alternative exon usage programs in neuronal development.
-
- Cell Biology
Amyotrophic lateral sclerosis (ALS) is a fatal neuromuscular disorder characterized by progressive weakness of almost all skeletal muscles, whereas extraocular muscles (EOMs) are comparatively spared. While hindlimb and diaphragm muscles of end-stage SOD1G93A (G93A) mice (a familial ALS mouse model) exhibit severe denervation and depletion of Pax7+satellite cells (SCs), we found that the pool of SCs and the integrity of neuromuscular junctions (NMJs) are maintained in EOMs. In cell sorting profiles, SCs derived from hindlimb and diaphragm muscles of G93A mice exhibit denervation-related activation, whereas SCs from EOMs of G93A mice display spontaneous (non-denervation-related) activation, similar to SCs from wild-type mice. Specifically, cultured EOM SCs contain more abundant transcripts of axon guidance molecules, including Cxcl12, along with more sustainable renewability than the diaphragm and hindlimb counterparts under differentiation pressure. In neuromuscular co-culture assays, AAV-delivery of Cxcl12 to G93A-hindlimb SC-derived myotubes enhances motor neuron axon extension and innervation, recapitulating the innervation capacity of EOM SC-derived myotubes. G93A mice fed with sodium butyrate (NaBu) supplementation exhibited less NMJ loss in hindlimb and diaphragm muscles. Additionally, SCs derived from G93A hindlimb and diaphragm muscles displayed elevated expression of Cxcl12 and improved renewability following NaBu treatment in vitro. Thus, the NaBu-induced transcriptomic changes resembling the patterns of EOM SCs may contribute to the beneficial effects observed in G93A mice. More broadly, the distinct transcriptomic profile of EOM SCs may offer novel therapeutic targets to slow progressive neuromuscular functional decay in ALS and provide possible ‘response biomarkers’ in pre-clinical and clinical studies.






