Molecular systematics of the subfamily Limenitidinae (Lepidoptera: Nymphalidae)
- Published
- Accepted
- Received
- Academic Editor
- Michael Wink
- Subject Areas
- Biodiversity, Entomology, Taxonomy
- Keywords
- Lepidoptera, Nymphalidae, Systematics, New tribe, Classification, Limenitidinae
- Copyright
- © 2018 Dhungel and Wahlberg
- Licence
- This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, reproduction and adaptation in any medium and for any purpose provided that it is properly attributed. For attribution, the original author(s), title, publication source (PeerJ) and either DOI or URL of the article must be cited.
- Cite this article
- 2018. Molecular systematics of the subfamily Limenitidinae (Lepidoptera: Nymphalidae) PeerJ 6:e4311 https://doi.org/10.7717/peerj.4311
Abstract
We studied the systematics of the subfamily Limenitidinae (Lepidoptera: Nymphalidae) using molecular methods to reconstruct a robust phylogenetic hypothesis. The molecular data matrix comprised 205 Limenitidinae species, four outgroups, and 11,327 aligned nucleotide sites using up to 18 genes per species of which seven genes (CycY, Exp1, Nex9, PolII, ProSup, PSb and UDPG6DH) have not previously been used in phylogenetic studies. We recovered the monophyly of the subfamily Limenitidinae and seven higher clades corresponding to four traditional tribes Parthenini, Adoliadini, Neptini, Limenitidini as well as three additional independent lineages. One contains the genera Harma + Cymothoe and likely a third, Bhagadatta, and the other two independent lineages lead to Pseudoneptis and to Pseudacraea. These independent lineages are circumscribed as new tribes. Parthenini was recovered as sister to rest of Limenitidinae, but the relationships of the remaining six lineages were ambiguous. A number of genera were found to be non-monophyletic, with Pantoporia, Euthalia, Athyma, and Parasarpa being polyphyletic, whereas Limenitis, Neptis, Bebearia, Euryphura, and Adelpha were paraphyletic.
Introduction
The butterfly family Nymphalidae has been the subject of intensive research in many fields of biology over the decades. However, the higher classification of the family is still being worked on, with the delineation of subfamilies being established fairly recently (Wahlberg et al., 2009). It is now clear that there are 12 subfamilies that are well supported by both molecular (Brower, 2000; Wahlberg, Weingartner & Nylin, 2003; Wahlberg et al., 2009) and morphological data (Freitas & Brown, 2004). These subfamilies have been accepted by most of the community working on Nymphalidae. The relationships of major lineages within subfamilies are now under scrutiny, with work at the level of subfamily already done on Apaturinae (Ohshima et al., 2010), Libytheinae (Kawahara, 2009), Nymphalinae (Wahlberg, Brower & Nylin, 2005) and Satyrinae (Peña et al., 2006), as well as a multitude of studies looking at relationships at lower levels within subfamilies. Here we turn our attention to Limenitidinae, a subfamily with a complex taxonomic history.
The rank and position of Limenitidinae has always been unstable and long debated among researchers. Popularly known as a “trash can” subfamily, Limenitidinae has included groups of species that could not be placed in any recognized subfamilies and were thus retained in the subfamily just for convenience (Harvey, 1991; Neild, 1996; Brower, 2000). Historically, Limenitidinae were placed as a tribe in the subfamily Nymphalinae (Smart, 1975). Later, Harvey (1991) placed Limenitidinae as the tribe Limenitidini in the subfamily Limenitidinae (sensu Harvey) but together with three unrelated tribes Coloburini (sensu Harvey), Biblidini (sensu Harvey), Cyrestidini (sensu Harvey), and two genera Pseudergolis and Stibochiona (now in the subfamily Pseudergolinae). Limenitidinae (sensu Harvey) is equivalent to Müller’s (1886) group III together with Cyrestidini (Harvey, 1991). Molecular work has finally unambiguously delineated the subfamily Limenitidinae (Brower, 2000; Wahlberg, Weingartner & Nylin, 2003; Wahlberg et al., 2009). Based on molecular data, the subfamily Limenitidinae is equivalent to the tribe Limenitidini of Harvey, it is sister to the subfamily Heliconiinae and does not include the taxa Cyrestidinae, Biblidinae, and Pseudergolinae (Brower, 2000; Wahlberg, Weingartner & Nylin, 2003; Freitas & Brown, 2004; Wahlberg et al., 2009).
As it is currently delineated, the subfamily Limenitidinae (Lepidoptera: Nymphalidae) comprises a little over 800 species placed in 46 genera and four tribes: Parthenini, Adoliadini (=Euthaliini), Limenitidini, and Neptini (Wahlberg, 2007). Limenitidinae are distributed worldwide and occur in all major biogeographical regions: Nearctic, Neotropics, Palaearctic, Afrotropics, Oriental, and Australasia (Chermock, 1950; Chou, 1998; Willmott, 2003). The species of the tribe Parthenini are limited to the Oriental and Australasian regions while the species of the tribes Neptini and Adoliadini are distributed throughout the Old World tropics. The species of the tribe Limenitidini are distributed mainly in the Palaearctic and the New World. It should be noted that some studies (e.g., Mullen et al., 2011) have included Lelecella as a limenitidine, although this genus is in fact in the subfamily Apaturinae.
Initial studies on Limenitidinae were mostly limited to the description of new species and genera. Schatz (1892) studied and classified the Limenitidinae of the world in three tribes (“Neptis-Gruppe”, “Limenitis-Gruppe”, and “Euthalia-Gruppe”) based on venation and palpal structures. Later, Reuter (1896) classified Limenitidinae into two tribes: Limenitidi and Neptidi based on studies of the palpi. The tribe Limenitidi (including the Euthalid complex) was further subdivided in two subtribes Limenitini and Parthenini. Moore (1890) surveyed the limenitidines of south eastern Asia introducing many new generic names and grouped them into two tribes Euthaliina and Limenitina (Neptis included) based on venation and maculation. Moore’s Euthaliina is a synonym of Adoliadina described earlier by Doubleday based on the genus Adolias (itself a synonym of Euthalia). Moore’s name Euthaliina has been in common use, as the following narrative shows. Aurivillius (1898) also surveyed and grouped the African Limenitidinae under two tribes Neptididi and Nymphalidi. According to Chermock (1950) most of species of Limenitidinae (except Neptis) can be distinguished from all other nymphalids by the first anal vein of the forewing that is preserved as a short spur at the base of the cubitus. Chermock (1950) considered Limenitidinae of the world to belong to one tribe Limenitini based on venation, male genitalia, life histories, maculation, palpal characters, and distribution. Based on egg morphology and following Eliot (1978) and Harvey (1991) divided the tribe Limenitidini into four subtribes: Limenitiditi, Neptiti, Partheniti, and Euthaliiti. However, Chou has divided Asian Limenitidinae into five tribes Euthaliini, Parthenini, Neptini, Limenitini, and Chalingini based on morphological characters (Chou, 1998; Zhang et al., 2011). Willmott (2003) suspected that Chalingini does not belong in Limenitidinae based on their unique morphology. In addition to ambiguous higher classification in Limenitidinae, many genera are vaguely defined or supported by few characters (Willmott, 2003).
The systematic relationships within Limenitidinae among its major lineages are still unclear. There have been some genus level phylogenetic studies (Willmott, 2003; Mullen, 2006; Mullen et al., 2011; Van Velzen et al., 2013; Ebel et al., 2015) and some phylogenetic studies included a few genera of the subfamily Limenitidinae (Zhang et al., 2008; Zhang et al., 2011; Wu et al., 2014). However, a comprehensive phylogenetic study of the entire subfamily at the genus and tribe level is still lacking, thus hindering evolutionary studies of the subfamily. Furthermore, a solid phylogenetic hypothesis of Limenitidinae is required to study the evolutionary processes that drive rates of diversification in the subfamily.
Our aims are to study systematics of the subfamily Limenitidinae using up to 18 gene regions per species of 205 taxa belonging to recognized genera and tribes of Limenitidinae spanning all major biogeographical areas. We also introduce seven new gene regions (CycY, Exp1, Nex9, PolII, ProSup, PSb and UDPG6DH) used in this study which have never been previously used for phylogenetic studies.
Material and methods
Taxon sampling
A total of 205 samples representing 39 genera and all four traditional tribes (Table S1): Parthenini, Neptini, Adoliadini and Limenitidini of the subfamily Limenitidinae were collected either by the authors during field visits or by various collaborators. Samples were acquired from all major biogeographical areas. Unfortunately, we could not obtain sequence data from three potentially important genera (Neurosigma, Euryphaedra, and Kumothales). Four exemplar taxa from the sister subfamily Heliconiinae: Argynnis, Heliconius, Actinote, and Cethosia were selected as outgroups to root the topology of the subfamily Limenitidinae.
Genomic DNA was mainly extracted from one or two legs, and in a few cases thoracic tissue, of dried mounted vouchers or ethanol-preserved specimens of butterflies. Genomic DNA was extracted using the Qiagen DNEasy extraction kit, following the protocol from the manufacturer. For each species, we amplified and sequenced one gene from mitochondrial genome (cytochrome oxidase subunit I, COI) and 17 genes from nuclear genomes, of which carbamoylphosphate synthetase (CAD), Ribosomal Protein S5 (RpS5), Ribosomal Protein S2 (Rps2), wingless (wgl), cytosolic malate dehydrogenase (MDH), glyceraldehydes-3-phosphate dehydrogenase (GAPDH), Elongation factor 1 alpha (EF-1a), Arginine Kinase (ArgKin), Isocitrate dehydrogenase (IDH) and dopa-decarboxylase (DDC) were amplified using primers and protocols from Wahlberg & Wheat (2008). For the new gene regions Cyclin Y (CycY), exportin-1-like (Exp1), sorting nexin-9-like (Nex9), DNA-directed RNA polymerase II polypeptide (PolII), suppressor of profiling 2 (ProSup), proteasome beta subunit (PSb), UDP glucose6 dehydrogenase (UDPG6DH) as well as a different section of ArgKin we used primer pairs and protocols described by Wahlberg et al. (2016). For a number of species, sequences were downloaded from GenBank (accession numbers in Table S1).
Successful amplicons were cleaned with A’SAP (ArticZymes) and Sanger sequenced (Macrogen Services, Amsterdam). Previously published DNA sequences (Wahlberg et al., 2009; Mullen et al., 2011; Van Velzen et al., 2013; Wu et al., 2014) were also included in the current study. Nucleotide sequence alignment was manually done using the program Bioedit (Hall, 1999). Sequences were managed and datasets were constructed using VoSeq v1.7.4 (Peña & Malm, 2012).
Phylogenetic inference
Phylogenetic analyses were done first separately for each gene (producing gene trees) and then for all the 18 genes combined. The combined dataset is given in Data S1. We explored various partitioning schemes of our concatenated multi-gene dataset using PartitionerFinder v1.1.1 (Lanfear et al., 2012) and compared them based on the Bayesian Information Criterion (BIC). We first partitioned by gene and codon positions and ran PartitionFinder in order to find which subsets could be combined. In addition, we calculated the relative rates of evolution for each site in the alignment using TIGER (Cummins & McInerney, 2011) and created partitions using the RatePartitions algorithm (Rota, Malm & Wahlberg, 2017). We tested a range of d values (2.0–5.0, with increments of 0.5), which affects the number of partitions, and calculated their BIC values in PartitionFinder.
Phylogenetic inference analyses were carried out using both Maximum likelihood (ML) and Bayesian Inference (BI) methods. Maximum likelihood phylogenetic inference analyses were carried out in RAxML v8.2.4 (Stamatakis, 2014) on XSEDE on the CIPRES Science Gateway v3.3 (Miller, Pfeiffer & Schwartz, 2010) using the best partition scheme suggested by the PartitionFinder/TIGER analysis based on BIC. For bootstrapping, we performed 1,000 Maximum Likelihood (ML) pseudo-replicates analyses and bootstrapping was performed under auto Majority Rule Criterion (autoMRE). Similarly, BI was performed using Markov Chain Monte Carlo (MCMC) in MrBayes v3.2.6 (Ronquist et al., 2012) on XSEDE on the CIPRES Science Gateway v3.3 (Miller, Pfeiffer & Schwartz, 2010). Two parallel runs of four chains (three heated and one cold) were performed for 20 million generations, with sampling done at every 1,000th generation. The software Tracer v1.6 (Rambaut et al., 2014) was used to inspect the sample sizes of the parameters used in the BI and also check for the convergence or otherwise of the parallel MCMC runs.
As there was a lot of missing data for many specimens (Table S1), we also analysed a subset of taxa that had 10 or more gene regions sequenced. This set of 55 taxa (including all the outgroups) was analysed with RAxML as described above, partitioned by gene.
Taxonomic decisions
The electronic version of this article in Portable Document Format (PDF) will represent a published work according to the International Commission on Zoological Nomenclature (ICZN), and hence the new names contained in the electronic version are effectively published under that Code from the electronic edition alone. This published work and the nomenclatural acts it contains have been registered in ZooBank, the online registration system for the ICZN. The ZooBank LSIDs (Life Science Identifiers) can be resolved and the associated information viewed through any standard web browser by appending the LSID to the prefix http://zoobank.org/. The LSID for this publication is: urn:lsid:zoobank.org:pub:A422503C-2E62-4001-8397-B8C9085CB23C. The online version of this work is archived and available from the following digital repositories: PeerJ, PubMed Central and CLOCKSS.
Results
Molecular data
Our final molecular data matrix consisted of 209 taxa representing 205 Limenitidinae species; four related taxa as outgroups; and 11,327 aligned nucleotide sites with no indels. In this study, we used 18 genes of which seven genes (CycY, Exp1, Nex9, PolII, Prosup, PSb and UDPG6DH) have not been previously used in phylogenetic studies of Nymphalidae butterflies. Table 1 gives the basic statistics for variation in each gene region. The new gene regions show similar amounts of variation to the standard gene regions of Wahlberg & Wheat (2008).
| Data set | Data type | Length (bp) | Dataset completion (%) | Variable (%) | Pars. Inf. (%) | Invariable (%) | Freq. A (%) | Freq. T/U (%) | Freq. C (%) | Freq. G (%) |
|---|---|---|---|---|---|---|---|---|---|---|
| ArgKin | Nuclear | 742 | 25.3 | 33.69 | 28.3 | 66.31 | 24.42 | 19.91 | 30.51 | 25.16 |
| CAD | Nuclear | 850 | 12.2 | 43.29 | 34.94 | 56.71 | 35.23 | 30.71 | 13.83 | 20.23 |
| COI | Mitochondrial | 1,475 | 84.6 | 49.97 | 40.54 | 50.03 | 29.15 | 40.03 | 16.16 | 14.66 |
| CycY | Nuclear | 375 | 24.8 | 36 | 31.47 | 64 | 31.83 | 31.78 | 15.69 | 20.7 |
| DDC | Nuclear | 373 | 14.4 | 44.24 | 38.34 | 55.76 | 25.4 | 28.96 | 24.38 | 21.25 |
| EF1a | Nuclear | 1,240 | 73.8 | 35.97 | 30.65 | 64.03 | 26.75 | 22.42 | 26.5 | 24.34 |
| Exp1 | Nuclear | 729 | 8.6 | 35.25 | 27.57 | 64.75 | 31.76 | 30.58 | 16.47 | 21.18 |
| GAPDH | Nuclear | 691 | 69.1 | 40.23 | 35.31 | 59.77 | 25.16 | 27.26 | 25.25 | 22.32 |
| IDH | Nuclear | 710 | 34.3 | 44.08 | 39.72 | 55.92 | 32.43 | 27.31 | 18.93 | 21.33 |
| MDH | Nuclear | 733 | 18.1 | 32.88 | 20.33 | 67.12 | 28.36 | 27.03 | 21.6 | 23.01 |
| Nex9 | Nuclear | 420 | 25 | 43.57 | 36.43 | 56.43 | 34.33 | 25.8 | 19.42 | 20.45 |
| PolII | Nuclear | 360 | 24.2 | 39.17 | 35.56 | 60.83 | 31.49 | 29.65 | 16.13 | 22.73 |
| ProSup | Nuclear | 432 | 14.1 | 39.35 | 29.4 | 60.65 | 26.84 | 30.72 | 18.46 | 23.99 |
| PSb | Nuclear | 366 | 24.1 | 42.62 | 40.16 | 57.38 | 28.86 | 26.14 | 22.31 | 22.68 |
| RpS2 | Nuclear | 411 | 23.9 | 39.42 | 34.06 | 60.58 | 24.98 | 24.59 | 21.68 | 28.75 |
| RpS5 | Nuclear | 617 | 63.5 | 41.82 | 38.74 | 58.18 | 27.44 | 25.19 | 23.01 | 24.35 |
| UDPG6DH | Nuclear | 405 | 16.9 | 37.78 | 35.8 | 62.22 | 30.15 | 28.79 | 19.9 | 21.17 |
| Wingless | Nuclear | 400 | 78.1 | 53.5 | 42.75 | 46.5 | 23.66 | 19.56 | 27.75 | 29.04 |
The best partitioning scheme was evaluated based on BIC values as calculated by PartitionFinder (Lanfear et al., 2012). Partitioning strategies based on genes were decisively worse than those based on RatePartitions or partitioning by gene and codon position (Table 2). The best partitioning scheme was created by RatePartitions with d = 5.0, which subdivided the data into 19 partitions. This partitioning scheme had a BIC value 543 units lower than the next best scheme based on partitioning by gene and codon position. We thus used the RatePartitions 5.0 scheme for further analyses.
| Partitions | BIC | Difference to best |
|---|---|---|
| Partition_18_genes | 334394.0202 | 12525.6029 |
| Partition_PF_gene | 334060.5349 | 12192.11755 |
| LimenTIG2.0_parts | 322668.1567 | 799.739362 |
| LimenTIG4.5_parts | 322615.2355 | 746.818151 |
| LimenTIG4.0_parts | 322546.4603 | 678.042952 |
| LimenTIG3.5_parts | 322518.0154 | 649.598091 |
| LimenTIG2.5_parts | 322517.0803 | 648.662997 |
| LimenTIG3.0_parts | 322498.6547 | 630.237363 |
| Partition_PF_codon | 322411.781 | 543.363707 |
| LimenTIG5.0_parts | 321868.4173 |
Systematics
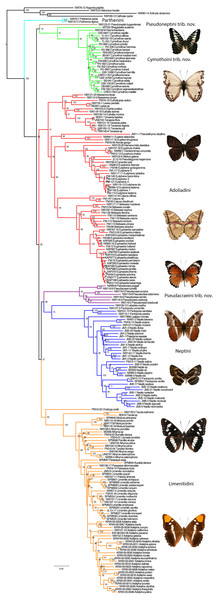
With four outgroups, the maximum Likelihood (ML) (Fig. 1) and Bayesian Inference (BI) (Fig. S1) methods recovered the subfamily Limenitidinae as monophyletic with strong bootstrap supports (BS 100) and high posterior probabilities (PP 1.0). Our analyses recovered seven major lineages: a clade including species of the tribe Parthenini, a clade including Bhagadatta, Harma and Cymothoe, a clade including Pseudacraea, a clade including species of the tribe Neptini, a clade including species of tribe Adoliadini, a clade including Chalinga pratti and species of the core tribe Limenitidini (Harvey, 1991) and finally an independent lineage leading to Pseudoneptis bugandensis of the tribe Limenitidini. Most of these clades are strongly supported, the exceptions are the position of Chalinga as sister to the core Limenitidini and the sister position of Bhagadatta to Cymothoe and Harma. The relationships of six of the seven lineages are not resolved despite increased gene region sampling, only the sister relationship of Parthenini to the rest of Limenitidinae is strongly supported. Reducing the taxon sampling to only those taxa with 10 or more gene regions sequenced did not change the fundamental results in any way (Fig. S2).
Figure 1: The Maximum Likelihood topology for Limenitidinae with associated bootstrap values.
Major lineages that are considered tribes in this paper are coloured. Examples of butterflies (voucher specimens for this work) from top: Parthenos sylvia, Cymothoe caenis, Euriphene tadema, Euphaedra herberti, Pseudacraea poggei, Lebadea martha, Neptis ida, Limenitis reducta and Adelpha californica.The relationships within the Cymothoe clade are very similar to those reported in a previous study of the genus (Van Velzen et al., 2013), with the exception of the genus Bhagadatta which appears to be sister to Cymothoe and Harma with low to moderate support (BS 57, PP 0.98).
The genus Pseudacraea formed an independent lineage that appears to be sister to Neptini with no support in ML (BS 39) and moderate support in BI (PP 0.98). Relationships of species within Pseudacraea were generally well supported and clear, with P. poggei and P. lucretis being the sister group of the rest of the genus.
In Neptini, we found the genus Lebadea to be sister to the rest of the tribe, with the core Pantoporia branching off next and Lasippa being sister to Neptis s.l. We recovered the genus Pantoporia as polyphyletic and Neptis as paraphyletic. The species Pantoporia venilia from Australia was found to be sister to a clade of African species of Neptis with low support values (BS 65, PP 0.53), but certainly within a strongly supported (BS 100, PP 1.0) derived clade of Neptis. The species Phaedyma aspasia was also found within one of the well supported Asian Neptis clades. Asian species of Neptis formed a grade while all sampled African species were found in a strongly supported clade (BS 98, PP 1.0).
In Adoliadini, we found five well supported clades, the Asian Euthalia clade, and the African Euptera, Hamanumida, Catuna and Bebearia clades. Relationships of these five lineages were not well supported, with the African clades forming a monophyletic group in the ML analyses (BS 65), but not in the BI analyses, where the Euptera clade was the sister to the rest of Adoliadini (PP 1.0). The Palaearctic species Abrota ganga was sister to the Hamanumida clade with very high support values (BS 99, PP 1.0) rather than clustering with other Asian Adoliadini. We recovered the genus Euthalia as polyphyletic, with Euthalia adonia being the sister to Dophla evelina with strong support while Euthalia monina was sister to species of the genera Tanaecia with strong support values. Bebearia was found to be paraphyletic with regard to Euphaedra with low bootstrap (BS 58) but high posterior probability (PP 0.99). The species Crenidomimas concordia was found to be nested within the species Euryphura chalcis with all three specimens being genetically very similar.
We found Chalinga pratti to be sister to the core Limenitidini with low or no support (BS 60, PP 0.56), but this position was consistent and stable in all analyses. Within the core Limenitidini there are five well supported lineages, with Tacola sister to the rest, Moduza branching off next, then the Athyma clade, and finally Parasarpa zayla as sister to the Limenitis clade. The genus Tarattia was found to be within Moduza. The Athyma clade comprises the paraphyletic Athyma with the genera Sumalia, Pandita and Lamasia deeply within the genus. Also the Limenitis clade shows nonmonophyletic genera: Parasarpa, Limenitis and Adelpha are intermixed and the clade contains the genera Auzakia and Patsuia. Relationships differ somewhat between the ML and BI analyses in this part of the tree, especially where branch lengths are very short or nonexistent.
Discussion
Systematic implications
Here, we studied molecular systematics of the recently defined (Wahlberg et al., 2009) subfamily Limenitidinae. Previous studies (Brower, 2000; Wahlberg, Weingartner & Nylin, 2003; Freitas & Brown, 2004) clearly showed that the traditional view of the subfamily Limenitidinae (e.g., Harvey, 1991) was not monophyletic. Wahlberg et al. (2009) defined the subfamily but did not discuss the internal relationships. We recovered seven independent lineages corresponding to four tribes Parthenini, Neptini, Adoliadini, Limenitidini; as well as three independent lineages without formal tribal names: the Cymothoe clade, Pseudoneptis and Pseudacraea (Fig. 1). For consistency, when discussing previous publications, we will align taxon concepts with ours, e.g., our concept of the subfamily Limenitidinae has often been referred to as the tribe Limenitidini, and our tribes as subtribes.
Many of the relationships we found were surprising, but some were anticipated by Willmott (2003) based on careful morphological comparisons. For instance he noted similarities in male genitalia between Lebadea and Neptis, suggested that Bhagadatta might be related to Cymothoe, that Tacola is sister to the rest of Limenitidini, maintained that Parthenos is the only genus to be included in Parthenini, and proposed that Cymothoe be placed in a tribe of its own. Willmott (2003) also suggested that Neptini was not a separate entity from Limenitidini, as did Amiet (2000), whereas based on our analyses it is clearly a separate entity that is not even sister to Limenitidini.
With the exception of the position of Parthenini as sister to the rest of Limenitidinae, the relationships of the major lineages within the subfamily were poorly supported despite up to 18 gene regions being sequenced for specimens within each lineage. The branches subtending these lineages are characterised by very short lengths, suggesting a period of rapid divergences. Such patterns are repeated throughout the evolutionary history of Limenitidinae, notably within Cymothoe (Van Velzen et al., 2013), Euriphene, Euphaedra and the base of the Limenitis clade.
Parthenini
As anticipated by Willmott (2003), our data recovered only species of Parthenos in this tribe and its position as sister to the rest of Limenitidinae was recovered with strong support in all phylogenetic analyses. Similar results were also found by Zhang et al. (2008), Zhang et al. (2011) and Wu et al. (2014). Parthenos is limited to the Indo-Australian region.
Cymothoini Dhungel & Wahlberg trib. nov.
LSID urn:lsid:zoobank.org:act:C26A6D77-EDE1-43DB-919F-254E47B82CA3
Based on our results, the genera Cymothoe, Harma and likely Bhagadatta form an independent lineage that warrant tribal status. Harvey (1991) classified the two African genera Harma and Cymothoe in the tribe Limenitidini. However, Amiet (2001) and Willmott (2003) regarded Cymothoe (including Harma) as incertae sedis, as they share more morphological features with Adoliadini than with Limenitidini. The genera Harma and Cymothoe were recovered as sister to each other with strong support values. The Harma + Cymothoe sister clade relationship was consistent with the previous study by Van Velzen et al. (2013). Harma and Cymothoe are here placed in a new tribe Cymothoini Dhungel & Wahlberg trib. nov. The tribe forms a strongly supported clade comprising species placed in Cymothoe and Harma with DNA sequence data from the following gene regions (exemplar sequences from Cymothoe caenis) ArgKin (GQ864537), CAD (GQ864636), COI (GQ864754), CycY (MG741765), DDC (MG741734), EF1a (GQ864848), GAPDH (GQ864952), IDH (GQ865083), MDH (GQ865196), Nex9 (MG741407), PolII (MG741353), ProSup (MG741316), PSb (MG741271), RpS2 (GQ865312), RpS5 (GQ865420), UDPG6DH (MG741133) and wingless (GQ864442).
Surprisingly, we recovered species Bhagadatta austenia as a sister to genera Harma + Cymothoe but with a weak support values (BS 57, PP 0.98). Bhagadatta austenia has been classified in the tribe Limenitidini by Harvey (1991) and Wu et al. (2014) but incertae sedis by Willmott (2003), who noted similarities in genitalia with Cymothoe. We retain Bhagadatta as incertae sedis in Limenitidinae, but suggest that it might be placed in the new tribe Cymothoini once further information is available. Interestingly, Bhagadatta is restricted to Asia whereas Harma and Cymothoe are African genera. Only COI sequences were available for Bhagadatta from the study of Wu et al. (2014), thus it is imperative that nuclear genes are sequenced from this taxon to test its position.
Neptini
Neptini including Lebadea was recovered as monophyletic with moderate support (BS 75, PP 0.98). The monotypic genus Lebadea was classified as a member of tribe Parthenini by Harvey, but Willmott (2003) removed it to Limenitidini and suggested similarities to Neptis in male genitalia. Wahlberg et al. (2009) found the genus to be sister to Neptini, with no comment, a position that we corroborate here with more data. The core Neptini, including the genera Neptis, Pantoporia, Lasippa and Phaedyma, form a strongly supported clade, with Pantoporia being sister to Lasippa and Neptis, and Phaedyma aspasia being within Neptis. Phaedyma aspasia was originally described in Neptis by Leech but has been placed in Phaedyma by various authors, e.g., Chou (1998). Unfortunately we were not able to sample the type species of the genus Phaedyma (P. heliodora, synonym of P. amphion), thus we are unable to say whether the genus should be synonymized with Neptis. We propose a revised combination, Neptis aspasia comb. rev. Similarly, Pantoporia venilia does not belong in the genus Pantoporia, but is clearly within Neptis, leading to another revised combination Neptis venilia comb. rev.
The species of Neptis are distributed throughout Asia, Africa, Australia, and Europe, with the center of diversity being SE Asia. Our results suggest that the African species form a monophyletic group, with four Asian clades forming a paraphyletic grade with regard to the African clade.
Pseudacraeini Dhungel & Wahlberg trib. nov.
LSID urn:lsid:zoobank.org:act:E9569B8F-4D9D-4BCC-A18F-431557043079
Our results recovered the genus Pseudacraea as a monophyletic group with strong support values, and suggest that Pseudacraea might be sister to Neptini, although with no support in ML. Pseudacraea has been classified as Limenitidini (Harvey, 1991; Willmott, 2003). Amiet (2000) and Willmott (2003) suggest that Pseudacraea share synapomorphies with Limenitidini and Neptini, and indeed our ML topology suggests that these three lineages form a monophyletic group, however with no support at all. It appears that Pseudacraea is an independent lineage much like Pseudoneptis and the Cymothoe clade, and is thus placed in a tribe of its own: Pseudacraeini Dhungel & Wahlberg trib. nov. The tribe comprises species in the genus Pseudacraea and can be characterized by the DNA sequence data from the following gene regions (example from Pseudacraea poggei) ArgKin (MG741852), CAD (GQ864704), COI (GQ864802), CycY (MG741798), EF1a (GQ864896), Exp1 (MG741609), GAPDH (GQ865024), IDH (GQ865143), MDH (GQ865258), Nex9 (MG741441), PolII (MG741387), ProSup (MG741336), PSb (MG741302), RpS2 (GQ865362), RpS5 (GQ865489), UDPG6DH (MG741157) and wingless (GQ864490).
Adoliadini
The monophyly of Adoliadini is strongly supported (BS 91, PP 1.0). This tribe contains species from genera that are distributed in both Asia and Africa. Based on biogeography, Adoliadini could be divided into two subtribes: Adoliadina (Euthalia clade) for the Asian and Bebearina (Hamanumida, Bebearia and Catuna clades) for African species. This division does not take into account the African Euptera clade, containing the genera Euptera and Pseudathyma, which does not have a stable position in our analyses, being either sister to all Adoliadini (BI, PP 1.0) or sister to the other African clades (ML, BS 65). This suggests that using the concept of subtribe is not particularly useful in this case. Surprisingly, the Asian genus Abrota was sister to the African Hamanumida clade with strong support values (BS 99, PP 1.0) rather than clustering with other Asian Adoliadini.
The genus Euthalia was recovered as paraphyletic with Euthalia monina being sister to Tanaecia and Euthalia adonia being sister to Dophla with strong support values. This pattern is intriguing and calls for a much more detailed study of the species rich genus Euthalia. Another intriguing pattern is the genetic similarity of Crenidomimas concordia with Euryphura chalcis. These two taxa are very different based on wing patterns, with Crenidomimas perhaps mimicking the genus Sevenia (Nymphalidae: Biblidinae), but clearly they are very closely related to each other and should be the focus of a more detailed study. The genus Bebearia was also found to be paraphyletic with regard to Euphaedra, although with only moderate support in ML analyses. This clade also requires further study in order to establish whether a new genus needs to be described.
Limenitidini
The position of Chalinga pratti (also known as Seokia pratti) as sister to the core Limenitidini was stable across all analyses, but never had high support. As noted in the Introduction, Chou (1998) placed Chalinga in its own tribe Chalingini and Willmott (2003) suspected that Chalinga (including Seokia) perhaps did not belong to Limenitidinae. Our results show that it does indeed belong to the subfamily, and is likely to be the sister group to the core Limenitidini. For the time being we prefer to keep Chalinga in the tribe Limenitidini until there is further evidence that it should be considered a separate lineage worthy of tribal status.
The core Limenitidini comprises five distinct lineages, of which three show para- and polyphyly of constituent genera. These are the Moduza, Athyma and Limenitis clades. In addition, the genus Tacola and the species Parasarpa zayla form independent lineages. Two species endemic to Sulawesi have been removed from Moduza and placed in the genus Tarattia (Hanafusa, 1989; Tsukada, 1991), of which we sampled T. lysania. We found T. lysania to be sister to Moduza lymire, also endemic to Sulawesi, but retained in the genus Moduza (Vane-Wright & De Jong, 2003). We suggest that until further evidence shows that the Sulawesian clade is clearly sister to Moduza and not within it, Tarattia should be considered a synonym of Moduza. The genus Athyma has three relatively small genera within it: Pandita, Sumalia, and Lamasia. Lamasia lyncides was separated from Moduza by Tsukada (1991), but appears to actually be a species of Athyma. As the three genera are well within Athyma, they should be synonymized with it.
The phylogenetic relationships of genera within the Limenitis clade are complex and unresolved. The type species of the genus Parasarpa (P. zayla) is an independent lineage sister to the Limenitis clade with good support, but other members of the genus are found within the clade in an unresolved position. Adelpha is found in two well supported clades that may or may not be sister to each other, a result also found by Mullen et al. (2011). The monotypic Patsuia appears to be sister to the type species of Limenitis (L. populi) and thus the former can be synonymized with the latter genus. The position of the monotypic Auzakia varies depending on the method of analysis, with ML placing it as sister to the rest of the Limenitis clade, while Bayesian inference places it within Limenitis. On the whole, the genus Limenitis presents a challenge for classification and clearly more data are necessary to resolve the relationships.
Pseudoneptini Dhungel & Wahlberg trib. nov.
LSID urn:lsid:zoobank.org:act:AB322712-F361-4FDD-A6C3-E9DEC6EA9402
The genus Pseudoneptis was classified in the tribe Limenitidini by Harvey (1991) but incertae sedis by Willmott (2003). In this study, Pseudoneptis is recovered as sister either to the Cymothoe clade or to Limenitidini depending on method of analysis, i.e., it is highly unstable. Given that we have sequenced 14 gene regions from our specimen, the instability is more likely to be due to a rapid divergence scenario than a lack of data. This suggests that Pseudoneptis should be placed in a tribe of its own, especially since the single species in the genus has a suite of apomorphies (Amiet, 2002). We thus erect a monotypic tribe Pseudoneptini Dhungel & Wahlberg trib. nov. for the species Pseudoneptis bugandensis. Apomorphies for the tribe are described in Amiet (2002) and the lineage is also diagnosed by the the unique combination of DNA sequence data from the following gene regions ArgKin (MG741830), CAD (GQ864705), COI (GQ864803), CycY (MG741777), EF1a (GQ864897), GAPDH (GQ865025), IDH (GQ865144), MDH (GQ865259), Nex9 (MG741419), PolII (MG741365), PSb (MG741283), RpS2 (GQ865363), UDPG6DH (MG741142) and wingless (GQ864491).
Conclusion
This study presents the most comprehensive phylogenetic analysis to date for the “trash-can” subfamily Limenitidinae. Based on fragments of up to 18 genes per species, 205 species and four outgroups, our results recovered Limenitidinae as a monophyletic clade and which comprises seven major lineages that deserve tribal status. Four tribes have been traditionally recognized: Parthenini, Neptini, Adoliadini, and Limenitidini, while three lineages are placed in new tribes here: Cymothoini, Pseudoneptini and Pseudacraeini. The new Cymothoini tribe includes two African genera Cymothoe and Harma, and quite likely an Asian genus Baghadatta. The latter two new tribes are monogeneric. At the genus level, we found several traditionally recognized genera to be either poly- or paraphyletic, i.e., Neptis, Euryphura, Pantoporia, Athyma, Parasarpa, Limenitis, and Adelpha. Further work increasing the taxon sampling is necessary to test the monophyly of these genera and revise their limits.
Supplemental Information
List of taxa used in this study with GenBank accession numbers for sequenced gene regions
The Bayesian Inference topology for Limenitidinae with associated Posterior Probabilty values
Major lineages that are considered tribes in this paper are coloured as in Fig. 1.
The Maximum Likelihood topology for Limenitidinae for taxa that have 10 or more gene regions sequenced
Values to the right of nodes are bootstrap values for that node. Major lineages that are considered tribes in this paper are coloured as in Fig. 1.