Molecular phylogeny and evolutionary history of Moricandia DC (Brassicaceae)
- Published
- Accepted
- Received
- Academic Editor
- Hugo Cerda
- Subject Areas
- Biodiversity, Evolutionary Studies, Taxonomy
- Keywords
- Molecular phylogeny, Moricandia, Brassicaceae, Phylogenetic tree, Dated tree, Eruca foleyi, Moricandia rytidocarpoides, Rytidocarpus, Biogeography
- Copyright
- © 2017 Perfectti et al.
- Licence
- This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, reproduction and adaptation in any medium and for any purpose provided that it is properly attributed. For attribution, the original author(s), title, publication source (PeerJ) and either DOI or URL of the article must be cited.
- Cite this article
- 2017. Molecular phylogeny and evolutionary history of Moricandia DC (Brassicaceae) PeerJ 5:e3964 https://doi.org/10.7717/peerj.3964
Abstract
Background
The phylogeny of tribe Brassiceae (Brassicaceae) has not yet been resolved because of its complex evolutionary history. This tribe comprises economically relevant species, including the genus Moricandia DC. This genus is currently distributed in North Africa, Middle East, Central Asia and Southern Europe, where it is associated with arid and semi-arid environments. Although some species of Moricandia have been used in several phylogenetic studies, the phylogeny of this genus is not well established.
Methods
Here we present a phylogenetic analysis of the genus Moricandia using a nuclear (the internal transcribed spacers of the ribosomal DNA) and two plastidial regions (parts of the NADH dehydrogenase subunit F gene and the trnT-trnF region). We also included in the analyses members of their sister genus Rytidocarpus and from the close genus Eruca.
Results
The phylogenetic analyses showed a clear and robust phylogeny of the genus Moricandia. The Bayesian inference tree was concordant with the maximum likelihood and timing trees, with the plastidial and nuclear trees showing only minor discrepancies. The genus Moricandia appears to be formed by two main lineages: the Iberian clade including three species, and the African clade including the four species inhabiting the Southern Mediterranean regions plus M. arvensis.
Discussion
We dated the main evolutionary events of this genus, showing that the origin of the Iberian clade probably occurred after a range expansion during the Messinian period, between 7.25 and 5.33 Ma. In that period, an extensive African-Iberian floral and faunal interchange occurred due to the existence of land bridges between Africa and Europa in what is, at present-days, the Strait of Gibraltar. We have demonstrated that a Spanish population previously ascribed to Rytidocarpus moricandioides is indeed a Moricandia species, and we propose to name it as M. rytidocarpoides sp. nov. In addition, in all the phylogenetic analyses, M. foleyi appeared outside the Moricandia lineage but within the genus Eruca. Therefore, M. foleyi should be excluded from the genus Moricandia and be ascribed, at least provisionally, to the genus Eruca.
Introduction
The Brassiceae tribe of the Brassicaceae family includes many economically important species that are useful as vegetables, edible oils, crop forages, condiments and fuel crops (Zelmer & McVetty, 2009). For this reason, this crucifer tribe has been the focus of a vast amount of genetic, agronomic, and ecological research (Gómez-Campo, 1999; Gupta, 2009; Kole, 2011; Schmidt & Bancroft, 2011). Despite this interest, the phylogenetic relationship between the Brassiceae species is not yet resolved, and consequently, several well-established Brassiceae genera, such as Brassica or Diplotaxis, are probably polyphyletic and artificial. One small but economically important genus in this tribe is Moricandia DC. This genus probably originated in the Mediterranean Basin (Pratap & Gupta, 2009) and is currently distributed in North Africa, Middle East, Central Asia and Southern Europe, where it is associated mostly with arid and semi-arid environments (Tahir & Watts, 2011). Some Moricandia species have being extensively studied because they show intermediate C3–C4 photosynthetic metabolism (Apel, Horstmann & Pfeffer, 1997; McVetty, Austin & Morgan, 1989), a feature that improves carbon assimilation and water use efficiency under drought conditions (Apel, Bauve & Ohle, 1984; McVetty, Austin & Morgan, 1989). This so-called Moricandia syndrome may have great agronomic interest since it could be transferred to Brassica species by hybridization, increasing crop yield under extreme climatic conditions and in marginal areas (Apel, Bauve & Ohle, 1984). Disentangling the phylogenetic relationship between Moricandia species is key to understanding how these traits have evolved and to determine the placement of this genus inside the Brassiceae tribe.
Moricandia individuals mainly show erect and branched stems with simple, exstipulate leaves, usually with entire or pinnated lobes (Gupta, 2009). Their flowers are actinomorphic-disymmetric and mostly of lilac color, although range from almost white to deep purple depending on the species and weather conditions. Their fruits are dehiscent two-valves siliques with one or two seed series per valve (Gupta, 2009). Moricandia shows a high variability in the morphological characters used for identification, making the taxonomy of this genus complex and controversial (Jiménez & Sánchez Gómez, 2012). Eight species are currently recognised in the genus Moricandia (Warwick & Sauder, 2005; Warwick, Francis & Al-Shehbaz, 2006; Tahir & Watts, 2011): (1) M. arvensis (L.) DC, (2) M. moricandioides (Boiss.) Heywood, (3) M. foetida Bourg. ex Coss., (4) M. suffruticosa (Desf.) Coss. & Durieu, (5) M. spinosa Pomel, (6) M. foleyi Batt., (7) M. sinaica Boiss., and (8) M. nitens (Viv.) Durieu & Barr (Sobrino Vesperinas, 1984; Warwick, Francis & Al-Shehbaz, 2006; Tahir & Watts, 2011). Moricandia arvensis is an annual to perennial herb widely distributed in the northwest Africa, Iberian Peninsula and southern Italy, from where it has even invaded other parts of the planet (De Bolós, 1946; Gómez-Campo, 1999). It is mostly a ruderal species associated to cultivated areas, roadsides and other human disturbed habitats. Moricandia moricandioides and M. foetida are two herbaceous species endemic to the Iberian Peninsula. The former is distributed in semi-arid environments of the eastern Spain whereas the latter is a narrow endemism inhabiting arid habitats of the southeast Spain (Sobrino Vesperinas, 1993). Moricandia suffruticosa and M. spinosa are suffruticose species inhabiting Morocco, Algeria, Tunisia and probably Libya, whereas M. foleyi is an annual herb showing a very narrow distribution in desert areas of southern Morocco and Algeria. Moricandia sinaica is a herbaceous species located in desert areas from the Near East to Pakistan; and M. nitens is a suffruticose species distributed from North Africa to Middle East. In addition, several subspecies and varieties have been named, in particular for the widely distributed M. arvensis (Schulz, 1936; Heywood, 1964; Maire, 1967).
The phylogenetic relationships among the Moricandia species, and of these with the rest of the Brassiceae species, have not been well established yet. Moricandia was previously included into the subtribe Moricandiinae (Schulz, 1923; Schulz, 1936; Gómez-Campo, 1980), although molecular evidences have shown little support for this subtribe (Warwick & Black, 1994; Warwick & Sauder, 2005). Most family-wide phylogenies have included some Moricandia species (e.g., Warwick & Black, 1997; Warwick & Sauder, 2005; Bailey et al., 2006; Jiménez & Sánchez Gómez, 2012; Schlüter et al., 2016), and these large-scale studies have suggested that Moricandia belongs to the Rapa/Oleracea subtribe together with Brassica, Diplotaxis, Enartrocarpus, Eruca, Erucastrum, Morisia, Raphanus, Rapistrum and Rytidocarpus (Warwick & Hall, 2009). Although Moricandia has been considered a Brassica coenospecies on the basis of cytogenetic and morphological similarities (Gómez-Campo, 1999), molecular phylogenetic analyses suggest that Moricandia is closely related to the genus Rytidocarpus and less clearly to Eruca (Warwick & Black, 1994; Warwick & Sauder, 2005; Bailey et al., 2006; Couvreur et al., 2010). The monotypic genus Rytidocarpus has long been recognized as very close to Moricandia because it presents Moricandia-like cotyledons, similar purple flowers and the same chromosome (2n = 28) complement (Gómez-Campo, 1980; Prakash et al., 2009). Thereby, due to this incomplete and sometimes contradictory evidence, the phylogenetic position and evolutionary history of Moricandia is still unresolved, despite the importance this information could have to understand the evolution of agronomic traits.
The main goal of this study is to disentangle the phylogenetic relationships among the Moricandia species, using a nuclear and two plastidial regions. In addition, we have dated the main events in the evolution of this genus and determined the phylogenetic relationship of Moricandia with its closely related genera Rytidocarpus and Eruca. We show that one Moricandia species should be excluded of this genus and demonstrate that a population previously ascribed to Rytidocarpus moricandioides is indeed a new Moricandia species.
| Taxon | Population code | Voucher | Location | Geographical coordinates |
|---|---|---|---|---|
| Moricandia arvensis | Mar01 | GDA62592 | Barranco del Espartal, Baza, Granada, Spain | 37°31′12″N 2°42′11.99″W |
| Mar33 | GDA62641 | Road between Santa Fe-La Malahá, Granada, Spain | 37°8′24″N 3°43′53.99″W | |
| Mar35 | MA321698-1 | Cortijo de las Monjas, Olula del Rio, Almería, Spain | 37°22′18″N 2°17′53.99″W | |
| Mar38 | MA50245-1 | Road to Mejorada del Campo, Madrid, Spain | 40°22′57.47″N 3°35′28.61″W | |
| Mar42 | MA321231-1 | Close to Road A-35, Mogente, Valencia, Spain | 38°52′17.22″N 0°46′23.52″W | |
| M. foetida | Mfo01 | GDA49837-1 | Road between Tabernas and Sorbas, Almería, Spain | 37°0′15.83″N 2°27′26.76″W |
| Mfo02 | GDA62639 | Olula del Rio, Almería, Spain | 37°22′18″N 2°17′53.99″W | |
| M. foleyi | Mfy01a | GDA62595 | Road between Rissani and Merzuga, Morocco | 31°16′53.99″N 4°16′30″W |
| Mfy02a | GDA62593 | Merzuga, Morocco | 31°3′30″N 4°0′42″W | |
| M. moricandioides baetica | Mmob01 | GDA 62596 | Barranco del Espartal, Baza, Granada, Spain | 37°31′12″N 2°42′11.99″W |
| M. moricandioides giennensis | Mmog06 | GDA62638 | Road between Quesada and Huesa, Jaén, Spain | 37°45′57.18″N 3°12′8.09″W |
| M. moricandioides moricandioides | Mmom05 | GDA62640 | Road between Santa Fe-La Malahá, Granada, Spain | 37°8′24″N 3°43′53.99″W |
| M. moricandioides pseudofoetida | Mmsf01 | MUB105856 | Near Puerto del Garruchal, Murcia, Spain | 38°6′59.99″N 1°21′0″W |
| M. moricandioides pseudofoetida | Mmsf02 | – | Pago del Barranco y de Chumilla, Murcia, Spain | 37°56′14″N 1°1′58″W |
| M. nitens | Mni03 | GDA62597 | Close to Agouim, Morocco | 31°10′7.2″N 7°29′15.72″W |
| M. spinosa | Mspi01 | GDA62598 | Road between Missour-Boulemane, Morocco | 33°2′8.63″N 4°4′4.98″W |
| M. suffruticosa | Msu01 | GDA62599 | Road between Taza and Aknour, Morocco | 3°23′50.16″N 3°54′24.77″W |
| Eruca pinnatifida (=E. vesicaria pinnatifida) | Erupinn01 | GDA62602 | Road between Missour and Boulemane, Morocco | 33°2′8.63″N 4°4′4.98″W |
| Erupinn02 | – | Merzuga, Morocco | 31°3′30″N 4°0′42″W | |
| E. vesicaria (=E. vesicaria vesicaria) | Eruves01 | GDA62643 | Barranco del Espartal, Baza, Granada, Spain | 37°31′12″N 2°42′11.99″W |
| Rytidocarpus moricandiodes | Rmorm01 | GDA62600 | Road Taza-Aknour, Morocco | 3°23′50.16″N 3°54′24.77″W |
| Rmorg02 | GDA62601 | Moulay Yacoub, Morocco | 34°7′27.12″N 5°12′14.87″W | |
| Rmorg01-13b | GDA62636 | Road A322, close to Quesada, Jaén, Spain | 37°50′36.73″N 3°8′22.17″W | |
| Rmorg01-14b | GDA62636 | Road A322, close to Quesada, Jaén, Spain | 37°50′36.73″N 3°8′22.17″W |
Materials and Methods
Taxon sampling
We collected leaf tissue of 1–5 individuals from a total of 17 populations of Moricandia (Table 1), including five populations of Moricandia arvensis, five populations of M. moricandioides (two of the subspecies pseudofoetida and one of each subspecies baetica, giennensis, and moricandioides), two populations of M. foetida, two of M. foleyi, and one of each M. nitens, M. suffruticosa and M. spinosa. In addition, we included two populations of Eruca pinnatifida and one of E. vesicaria. Four populations of Rytidocarpus moricandioides, two from Morocco and one from Spain (sampled two consecutive years), were also included as representatives of this monotypic genus, which is probably the sister genus of Moricandia (Warwick & Sauder, 2005). These samples constituted the 24-samples set. Table 1 shows the code and location of these populations as well as the reference voucher material.
We also downloaded all the Moricandia-related ITS sequences hosted in GenBank (downloaded on March 1st, 2016). After quality-checking, we discarded those sequences that did not show complete ITS1 and ITS2 sequences and finally kept 18 Moricandia, 4 Eruca and 2 Rytidocarpus sequences for the following analyses. Specifically, we included seven M. arvensis (AY722472, DQ249832, EF601897, EF601898, EF601899, EF601900—var. robusta-, EF601901—var. garamantum-), one M. foetida (EF601902), one M. foleyi (EF601903), three M. moricandioides (AY722473, KF849875, EF601904), two M. nitens (AY722474, EF601905), one M. sinaica (EF601906), one M. spinosa (EF601907), two M. suffruticosa (AY722475, EF601908), two R. moricandioides (AY722483, EF601910), two E. vesicaria sativa (AY254536, DQ249821), one E. vesicaria vesicaria (AY722459), and one E. pinnatifida (AY722458) accessions.
DNA extraction, PCR amplification, and sequencing
Leaf tissues were freshly sampled from the specimens and subsequently desiccated and preserved in silica gel until DNA extraction. For each individual sample at least 60 mg of plant material was disrupted with a Mixer Mill MM400 (Retsch, Haan, Germany) using 2 mm steel beads. DNA was extracted using the GenElute Plant Genomic DNA Miniprep Kit (Sigma-Aldrich, St. Louis, MO, USA) following manufacturer’s instructions.
One nuclear and two chloroplast DNA regions were amplified and sequenced. The nuclear sequence was composed by the internal transcribed spacers of the ribosomal DNA (ITS1 and ITS2) and the 5.8 rDNA between both ITSs sequences, together with partial 18S and 28S sequences, with jointly spans ∼700 base pairs. The plastidial regions span 2004 base pairs for the NADH dehydrogenase subunit F (ndhF) gene and ∼1,600 base pairs for the trnT-trnF region, including intergenic spacers.
ITS regions were amplified with primers ITS1, ITS2, ITS3 and ITS4 (White et al., 1990) anchoring to ribosomal flanking regions using the following PCR conditions: 3 min at 94 °C as initial denaturing step, followed by 35 cycles of 15 s at 94 °C, 30 s at 64 °C (ITS1–ITS2 primers) or 53 °C (ITS3–ITS4 primers) and 45 s at 72 °C, and a final step of 3 min at 72 °C. The ndhF region was amplified using primers ndhF5, ndhF599, ndhF1354, and ndhF2100 (Taberlet et al., 1991) and the following PCR conditions: 3 min at 94 °C as initial denaturing step, followed by 35 cycles of 15 s at 94 °C, 30 s at 47 °C (for both primer pairs: ndhF5–ndhF1354 and ndhF599–ndhF2100) and 90 s at 72 °C, and a final step of 3 min at 72 °C. The trnT-trnF region was amplified with primers tabA, tabD, tabC and tabF (Taberlet et al., 1991). The PCR conditions for these regions were 3 min at 94 °C as initial denaturing step, followed by 35 cycles of 15 s at 94 °C, 30 s at 53 °C (tabA–tabD primers) or 58 °C (tabC–tabF primers) and 90 s at 72 °C, and a final step of 3 min at 72 °C. All PCR reactions were performed in an Eppendorf™ S Mastercycler (Eppendorf, Hamburg, Germany). Amplicons were precipitated by centrifugation at 4 °C after the addition of 0.15 volumes of 3 M sodium acetate, pH 4.6, and 3 volumes of 95% (v/v) ethanol. Amplicons were sent to Macrogen Europe (Geumchun-gu, Seoul, Korea) for sequencing in both directions, using their corresponding PCR primers. Chromatograms were reviewed and contigs were produced using Geneious v. 9 (Kearse et al., 2012; Biomatters, Inc., San Francisco, CA, USA, http://www.geneious.com) and thorough revised and corrected by eye inspection. Sequences were uploaded to GenBank (accession numbers in Table S1).
Sequence alignment and phylogenetic analysis
For the phylogenetic analyses we built two sequence sets: (1) the 24-samples set including the ITS, ndhF and trnT-trnF sequences for the sampled specimens, and (2) the GenBank-ITS set including the ITS sequences of the 24-samples set plus the 24 complete ITS sequences obtained from GenBank. We used Brassica rapa and Raphanus sativus sequences obtained from GenBank as roots. These genera belong to the same Rapa/Oleraceae lineage inside the tribe Brassiceae as Eruca, Rytidocarpus and Moricandia (Warwick & Hall, 2009). We used accessions KM538956 (B. rapa) and AY746462 (R. sativus) for ITS, and extracted the plastidial ndhF and trnT-trnF regions from the complete cpDNA accessions of both species (DQ231548 for B. rapa and K5716483 for R. sativus).
Sequences were aligned and concatenated with Geneious v.9 (Biomatters Ltd.) using the matff algorithm (Katoh et al., 2002) with posterior slight manual adjustments. Ribosomic 5.8S, and 18S and 28S flanking sequences did not show variation and were not included in the following procedures. Evolutionary substitution models were separately fitted for each DNA region using jModeltest 2.1.7 (Darriba et al., 2012). The best-fitted molecular evolutionary models under Bayesian Information Criterion were TPM3+G for ITS1, TIM2ef+G for ITS2, TPM1uf+I for ndhF, and TPM1uf+G for the trnT-trnF.
We reconstructed the phylogeny under Bayesian inference using MrBayes 3.2.1 (Ronquist et al., 2012). Two independent runs of six MCMC chains were run for 2 × 106 generations, sampling trees every 100 generations. Evolutionary models were implemented as GTR+G for ITS1, ITS2 and trnT-trnF, and as GTR with a proportion of invariable sites for ndhF. We checked convergence using Tracer v1.6.1 (Rambaut et al., 2014) and discarded, as the burn-in phase, the first 20% of the saved trees. The consensus tree was obtained with the remained trees. This process was performed with the 24-samples set for the entire sequence concatenation, for only ITS sequences and for only cpDNA sequences (ndhF and trnT-trnF). In addition, we run MrBayes with the GenBank-ITS-set using the same parameter setting.
To compare the obtained Bayesian inference tree with other possible tree topologies, we performed Bayes factor analysis (Kass & Raftery, 1995), using MrBayes 3.2.1 to calculate the marginal likelihoods (estimated using stepping-stone sampling based on 50 steps with 39,000 generations (78 samples) within each step).
We used the RAxML software (Stamatakis, 2014) for maximum likelihood phylogenetic inference of the 24-samples set, using a partition model (ITS1, ITS2, ndhF and trnT-trnF) assuming GTR+G substitution models. We executed 100 initial fast parsimony inferences and thereafter a thorough ML search. For confidence analysis, a bootstrap of 100 replicates was also performed.
We checked whether the evolution of Moricandia was congruent with a molecular clock by using a likelihood ratio test as implemented in MEGA v.7 (Kumar, Stecher & Tamura, 2015). This test compares the ML values of the Maximum Likelihood tree obtained with and without assuming a molecular clock under a GTR evolutionary model.
To produce a dated chronogram we used Beast 2.0 (Bouckaert et al., 2014) following two approaches. First, we used the substitution rate for non-codifying plastidial DNA obtained from the literature (1.2–1.7 × 109 substitution/site/year; Graur & Li, 2000). Second, since the Iberian species appeared as monophyletic (see results), we used the Messinian Salinity Crisis (5.9 Ma) and the Zanclean Flood (5.33 Ma) as a calibration uniform interval for the separation of the Iberian species from the rest of the Moricandia genus. To obtain a more inclusive tree with all the Moricandia species, we included the M. sinaica ITS sequence obtained from GenBank in this approach. The Bayesian search for tree topologies and node ages were conducted during 20,000,000 generations in BEAST using the previously fitted substitution models and using a strict clock and a Yule process as priors. MCMC were sampled every 1,000 generations and used a burn-in of 10%. Appropriate sampling in the stationary phase was checked using Tracer v1.6.1 (Rambaut et al., 2014).
The electronic version of this article in Portable Document Format (PDF) will represent a published work according to the International Code of Nomenclature for algae, fungi, and plants (ICN), and hence the new names contained in the electronic version are effectively published under that Code from the electronic edition alone. In addition, new names contained in this work which have been issued with identifiers by IPNI will eventually be made available to the Global Names Index. The IPNI LSIDs can be resolved and the associated information viewed through any standard web browser by appending the LSID contained in this publication to the prefix “http://ipni.org/”. The online version of this work is archived and available from the following digital repositories: PeerJ, PubMed Central, and CLOCKSS.
Results
Ribosomic 5.8S, and 18S and 28S flanking sequences did not show variation and were not included in the final alignment of the 24-samples set and the two outgroups, that spanned 4,055 bp. ITS1 extended over 274 bp and included 63 variable sites with 51 parsimonious informative positions. ITS2 spanned 194 bp and included 29 variable sites with 22 parsimonious informative positions. The amplified ndhF region spanned 2,004 bp with 87 variable sites and 58 parsimonious informative positions. No indels or stop codons were found on this region, which codifies for part of the NADH dehydrogenase subunit F. The trnT-trnF amplified region spanned 1,583 bp with 264 variable sites and 186 parsimonious informative positions.
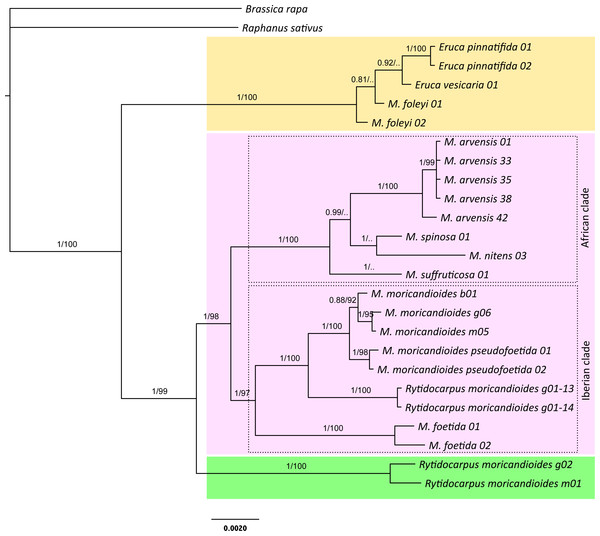
Figure 1: Bayesian inference tree produced with the complete 24-samples set.
In different color boxes are shown the genus Eruca (yellow), Moricandia (purple), and Rytidocarpus (green). The Iberian and African Moricandia clades are shown in doted line boxes. Branch labels are posterior probability values and bootstrap values obtained from the maximum likelihood phylogeny (Fig. S1). Scale in estimated substitutions per site. Note Rytidocarpus moricandioides samples g01-13 and g01-14 are proposed as the new species Moricandia rytidocarpoides.Join nuclear-plastidial phylogenetic trees
The Bayesian inference phylogeny for the complete 24-samples set, based on the nuclear and the plastidial regions, is shown in Fig. 1. The average standard deviation of split frequencies was 0.002 for the last generation, all estimated sample sizes were high (ESS > 960), and the potential scale reduction factor (PSRF) was 1 for all the parameters, indicating good MCMC mixing and sampling. Convergence metrics clearly indicated that MCMC converged to a well-supported topology. In fact, posterior probabilities showed high values for branches separating the different species. In this tree, the genus Moricandia did not appear as a monophyletic clade. The two samples of the endemic M. foleyi were arranged into a clade also containing the samples from Eruca and showing a posterior probability of 1. We performed a Bayes factor analysis to compare this topology (H1: Eruca and M. foleyi forming a monophyletic clade) to those constraining Eruca and M. foleyi to different clades (H2). The marginal likelihoods (in natural log units) estimated with MrBayes using stepping-stone sampling were −7987.71 for H1 and −8113.81 for H2. This difference implies that H1 is strongly supported by the Bayes factor test. Therefore, the taxonomic status of M. foleyi should be reconsidered.
The rest of the Moricandia species formed two clades. The first one includes the five M. arvensis samples, together with M. spinosa, M. nitens and, more basal, M. suffruticosa. We called this clade as the ‘African clade’ because all these species inhabit in Africa. The second clade—the ‘Iberian clade’—included the Spanish endemic species M. moricandioides and M. foetida, and the Rytidocarpus samples from Spain. The two samples belonging to subspecies M. moricandioides pseudofoetida appeared inside the branch of M. moricandioides but separated from the other M. moricandioides samples.
The genus Rytidocarpus appeared as the sister genus of Moricandia. The R. moricandioides samples from Morocco (Rmorg02-14-01 from Moulay Yacoub, close to Fez, and Rmorm01-14-01 collected in the road between the cities of Taza and Aknour; Table S1) form a monophyletic group. However, the R. moricandioides collected in Jaén, Spain, appeared inside the Moricandia Iberian clade, as a sister species of M. moricandioides (Fig. 1). To test the confidence of this result, we performed a Bayes factor analysis to test the topological hypothesis that the Spanish samples of Rytidocarpus are more closely related to Moricandia than to the other Rytidocarpus (Moroccan samples). Specifically, we compared the hypothesis that Rytidocarpus form a monophyletic group (H1; i.e., these Spanish samples are bona-fide Rytidocarpus) with the hypothesis that the two origins (Morocco and Spain) represent two different clades (H2). The marginal likelihoods (in natural log units) estimated with MrBayes using stepping-stone sampling were −8037.09 for H1 and −7997.49 for H2. This difference is very strong (decisive in the sense of Kass & Raftery, 1995) in favor of H2, and implies that Spanish sample should not be ascribed to Rytidocarpus but be recognized as a new species: M. rytidocarpoides (see Appendix for a formal description).
The ML tree produced with the 24-samples set (Fig. S1) was congruent with the Bayesian tree. They differed in the position of M. suffruticosa that appeared as the most basal species of the African clade, but with a low branch support. In addition, the position of M. spinosa and M. nitens also slightly varied with respect to the Bayesian tree, but these branching events were not supported with high bootstrap values (Fig. S1).
Plastidial and nuclear phylogenetic trees
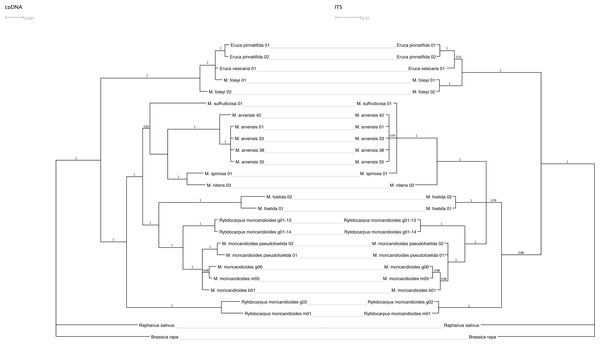
Figure 2 shows the Bayesian inference tree obtained with the combined cpDNA regions (ndhF and trnT-trnF) confronted with the tree obtained with the nuclear ITS regions using the same inference approach. The general pattern was maintained except for a lower resolution in the nuclear tree, where the Moricandia lineage appeared not so well resolved showing a basal trichotomy, and for minor rearrangements. For instance, M. spinosa and M. nitens appeared as sibling species in the plastidial tree but they were less closely related in the nuclear tree, although in both cases they belonged to the same African clade. It is also noticeable that M. foleyi samples appeared as monophyletic in the nuclear tree but interspersed with the Eruca samples in the plastidial tree. These trees were also obtained by ML inference and showed the same topologies (Fig. S2).
Figure 2: Tanglegram showing the Bayesian inference trees obtained from cpDNA (ndhF+trnT-trnF) and nuclear (ITS1+ITS2) sequences.
Branch labels refer to posterior probabilities. Scale in estimated substitutions per site.Figure 3: Phylogenetic tree based on ITS sequences from the ITS-set.
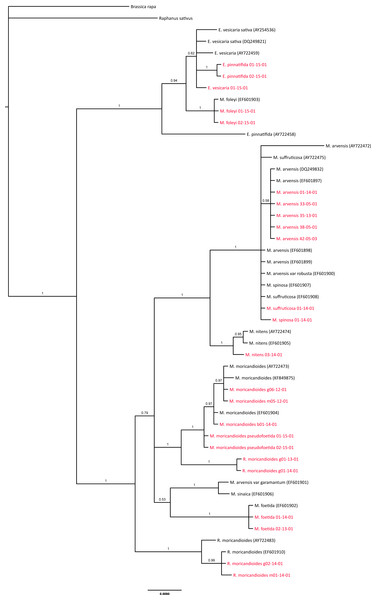
OTUs also included in the 24-samples set are depicted in red. Branch labels refer to posterior probabilities. Genbank accession codes are depicted between parentheses. Scale in estimated substitutions per site.ITS inclusive phylogeny
As a cross-validation of our sampling and to infer the phylogenetic position of M. sinaica, which is the only species of this genus that we could not sample, we used all complete ITS sequences available in GenBank to produce a comprehensive ITS phylogenetic tree (Fig. 3). The general pattern was compatible with our previous analyses. All our Moricandia samples were located close to GenBank ITS sequences of the same species. However, the ITS tree showed lower resolution than the combined nuclear-plastidial tree. In the ITS tree, Moricandia showed a basal polytomy formed by four lineages. The first one included the M. arvensis, M. suffruticosa and M. spinosa accessions. The second lineage included the M. moricandioides accessions and the R. moricandioides sampled in Jaén, Spain. The third and fourth lineages appeared in a clade without enough support (posterior probability = 0.53) to be considered as a unique lineage. Therefore, the third lineage was formed by M. sinaica and M. arvensis var. garamantum, and the fourth was composed by the M. foetida samples. Again, R. moricandioides appeared as the sister genus of Moricandia and M. foleyi accessions were grouped together with Eruca accessions. The E. pinnatifida accession AY722458 showed a distinctively long branch, probably indicating sequencing errors.
Chronogram
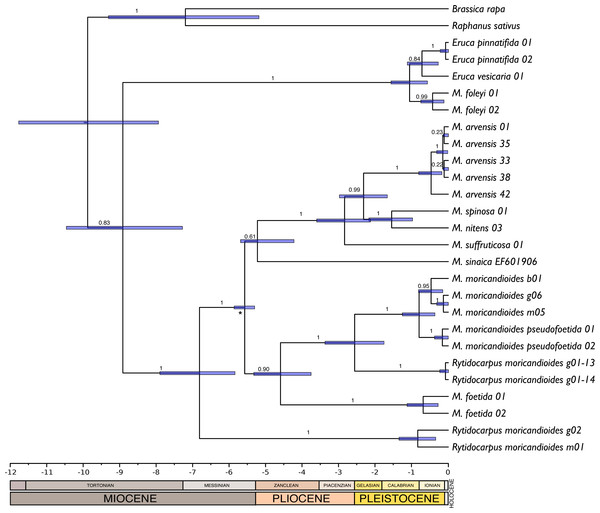
There were no significant differences between the trees obtained with or without assuming a clock (P = 0.92; LnL = −7851.663 enforcing a clock; LnL = −7835.925 without assuming a clock). Therefore, the null hypothesis of equal evolutionary rates (i.e., molecular clock) was not rejected and these sequences can be assumed to follow a global molecular clock. With this assumption, we inferred chronograms using a constant rate of 1.2 × 109 substitution/site/year for the non-codifying plastidial DNA. The dating of the monophyletic Iberian clade was compatible with an origin during the end of the Messinian period (5.99–5.33 Ma). Other trees obtained with substitution rates reported in the literature (1.2–1.7 × 109 s/s/y) were also compatible with this temporal window. Following, we produced a new chronogram constraining the origin of the Iberian clade to the Messinian salinity crisis period and including the ITS sequence of M. sinaica obtained from GenBank. This tree (Fig. 4) was congruent with that obtained with MrBayes (Fig. 1). The only topological differences were that the two samples of M. foleyi appeared to form a clearer lineage within the genus Eruca, and the position of M. sinaica, which was not included in the 24-samples set. In this chronogram the genus Moricandia separated 6.81 M years ago [5.83–7.89 Ma] from Rytidocarpus.
Figure 4: Chronogram obtained by Bayesian inference with the complete 24-samples set plus M. sinaica.
Branch labels refer to posterior probabilities. An asterisk marks the internal node used for calibration. Bars represent 95% confidence intervals. Temporal scale in Ma (million years ago).Discussion
The phylogenetic analyses presented here represent the evolution of the Moricandia genus clearly. Not only the Bayesian inference tree was concordant with the ML and timing trees, but also the cpDNA and ITS trees showed a fair agreement, with only minor discrepancies. In addition, all our samples were congruent with the GenBank accessions that were included in the ITS inclusive tree, although the latter tree showed lower resolution. In the trees of highest confidence, Moricandia appeared to be formed by two main lineages: the Iberian clade containing the Iberian species M. moricandioides, M. foetida and the new M. rytidocarpoides; and the African clade containing the species inhabiting the Southern Mediterranean region (M. sinaica, M. suffruticosa, M. nitens, M. spinosa and M. arvensis). The Bayesian inference trees showed high branch supports for this branching pattern and a good agreement with a molecular clock-like evolution. The timing of 7.19 Ma [5.18–9.23] for the separation of B. rapa and R. sativus, the two outgroup taxa in the chronogram, is congruent with the data previously reported (∼5.17 ± 2.5 Ma; Hohmann et al., 2015). This congruence is noteworthy since we used a strict clock that imposed a similar evolutionary rate in all lineages, which might not be realistic for the more distant (outgroup) taxa (Drummond et al., 2006). Therefore, we think that these phylogenetic trees are congruent and robustly represent the evolution of these taxa.
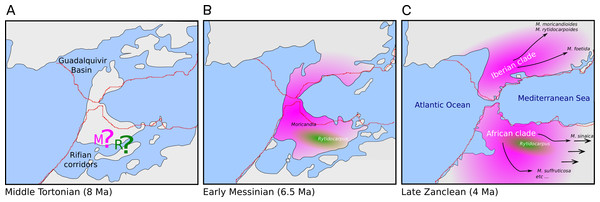
The genus Moricandia probably has a North African origin. Seven out of the previously eight recognized Moricandia species form a monophyletic group originated 6.81 Ma [5.83–7.89 Ma] after the separation from the Rytidocarpus lineage, their sister genera. Since Rytidocarpus is a North African endemism (Maire, 1967) and the genus Moricandia show a high number of species inhabiting North Africa (Maire, 1967), we support that Moricandia originated in North Africa, as was previously suggested (Sobrino Vesperinas, 1978). The colonization of the Iberian Peninsula probably occurred after a range expansion during the Messinian period, between 7.25 and 5.33 Ma (Fig. 5), coinciding with extensive African-Iberian floral and faunal interchanges (e.g., Fernández-Mazuecos & Vargas, 2011; Gibert et al., 2013). In that period, land bridges between North Africa and South Iberia appeared due to tectonic uplift coinciding with the isolation of the Mediterranean Sea from the Atlantic Ocean circa 6.3 Ma (Martín, Braga & Betzler, 2001). After the new aperture of the Strait of Gibraltar during the Zanclean period, the Iberian and African clades were separated and began to diverge (Figs. 4 and 5). A similar biogeographical pattern has been found for other plants. For instance, the genus Antirrhinum produced several lineages separated by the Straight of Gibraltar (Vargas et al., 2009), as well as the genus Hedera and the Saxifraga globulifera–reuteriana complex (Vargas et al., 1999; Vargas, Morton & Jury, 1999). However, this is not a general biogeographical pattern since other species have maintained genetic connections along the two sides of the Straight of Gibraltar (Rodríguez-Sánchez et al., 2008).
Figure 5: Hypothesis of the biogeography of the genus Moricandia coupled to the geological events at the end of the Miocene (A: Middle Tortonian; B: Early Messinian) and early Pliocene (C: Late Zanclean) in the Betic-Rifean Arch, the Strait of Gibraltar at current times.
Red lines depict the coastal lines at the present time. Based on the paleogeographical reconstruction of Martín et al. (2009).Moricandia sinaica is the most basal taxon of the African clade, arising circa 5.22 Ma (Fig. 4), although, given the moderate support of this branching event, its phylogenetic position should be accepted with cautiousness. In the inclusive ITS tree, which included the GenBank-ITS set plus the 24-samples set, M. sinaica (inhabiting Egypt and West Asia) and M. arvensis var. garamantum (from South Algeria) appeared as close relatives. This fact suggests that these two taxa could be the same species, as it has been previously postulated (Sobrino Vesperinas, 1984). The rest of the species forming the African clade (M. suffruticosa, M. nitens, M. spinosa and M. arvensis) can easily hybridize (Sobrino Vesperinas, 1997) and have been included in the same cytodeme (Prakash et al., 2009). Sobrino Vesperinas (1984) postulated that the now widely distributed M. arvensis is the same as the M. arvensis var. robusta from the Constantine area in Algeria (Gómez-Campo, 1999). Unfortunately, the ITS sequences included here did not show enough resolution to support or deny this claim. In the inclusive ITS tree (Fig. 3), M. arvensis var. robusta appeared in the same clade as the other M. arvensis samples, but also forming a polytomy with M. suffruticosa and M. spinosa. These two latter species show caryotypes with higher ploidy than the typical 2n = 28 (Sobrino Vesperinas, 1984) of this genus (namely, M. suffruticosa with 2n = 56 and M. spinosa with 2n = 84; Harberd, 1976), but in the same ploidy series than the other Moricandia species.
The caryotype of M. spinosa suggests that this species could be a stabilized hybrid. We have found that M. spinosa shows a cpDNA closely related to those of M. nitens but for the nuclear ITS markers shows a more distant genetic relationship (Fig. 2). For these last markers, M. spinosa is more similar to M. suffruticosa (Fig. 2). These facts suggest that M. spinosa (2n = 84) is an amphidiploid species produced by whole genome duplication after a hybridization event between M. suffruticosa (2n = 56) and M. nitens (2n = 28). These kind of events have been pervasive in the evolution of this tribe and family (Marhold & Lihová, 2006; Franzke et al., 2011). However, more genetic analyses are necessary to confirm this hypothesis.
After the Zanclean aperture of the Strait of Gibraltar, the Iberian Moricandia clade diverged into three well-supported lineages that can be now identified as three genuine species (Figs. 4 and 5). Of these species, the endemic M. foetida was the first to diverge and is now completely reproductively isolated from M. moricandioides. In fact, Sobrino Vesperinas (1993, 1997) experimentally demonstrated that they could not hybridize. The second lineage includes all the M. moricandioides samples in a monophyletic cluster. Five subspecies have been identified for this species based on morphological characters (Sobrino Vesperinas, 1993; Sánchez Gómez et al., 2001), with subspecies pseudofoetida showing intermediate characteristics with M. foetida (Sánchez Gómez et al., 2001). Jiménez & Sánchez Gómez (2012) based on ISSR markers proposed that M. moricandia pseudofoetida arose by reproductive isolation rather than hybridization between M. foetida and M. moricandioides moricandioides. Our current results do not refute that hypothesis and show that this subspecies, that inhabit similar environments that M. foetida, is clearly a M. moricandioides subspecies without showing signal of past hybridization.
The third species in the Iberian clade are plants growing in marls and calciferous substrates in a few places of the Guadiana Menor River Basin. Based mostly on fruit morphology, these plants were originally ascribed to the Moroccan-endemism Rytidocarpus moricandioides by Morillas-Sánchez & Fernández López (1995), and later accepted in the Vascular Flora of Eastern Andalusia (Blanca et al., 2011). These plants differ from other Moricandia in some peculiar morphological characters. Namely, they present sepals with scarious margins that persist during fruit development and fruits with two segments, the upper with beak shape, being similar to Rytidocarpus, whereas the other Moricandia species present non-persistent sepals without scarious margins and fruit in a siliqua. However, despite these morphological differences, our phylogenetic study clearly shows that they belong to the genus Moricandia (Figs. 1–4), inside the Iberian clade as a sister species to M. moricandioides. In addition, an exhaustive morphological comparison between the Jaén and the Morocco specimens clearly separated the Spanish samples from the genuine, Moroccan, Rytidocarpus moricandioides (see Tables S2, S3 and Figs. S3 S4). Therefore, these plants should be ascribed to the genus Moricandia and they deserve the taxonomic rank of species due to both their phylogenetic position and their distinctive morphological traits. We proposed the name Moricandia rytidocarpoides to denominate this new species Moricandia rytidocarpoides Lorite, Perfectti, Gómez, González-Megías & Abdelaziz sp.nov. urn:lsid:ipni.org:names:77166015-1 (see Appendix for a formal taxonomical description). Consequently, the siliqua fruits and the dehiscent scarious sepals are no longer defining characteristics (i.e., diagnostic traits) of the genus Moricandia. Several molecular analyses have demonstrated that fruit morphology shows homoplasy in Brassicaceae (Appel & Al-Shehbaz, 2003; Koch, Al-Shehbaz & Mummenhoff, 2003), and, therefore, fruit traits are not good indicators of phylogenetic relationships despite they have been widely used as taxonomic diagnostic traits (Hall et al., 2006).
The taxonomic status of Moricandia foleyi should be also amended. In all the phylogenetic analyses performed here M. foleyi appeared outside the Moricandia lineage and within the lineage of the Eruca species. Eruca is a genus of annual, non-hetero-arthrocarpic plants with a controversial taxonomy. Depending on the taxonomical treatment, Eruca includes from one to four species. Gómez-Campo (1999) considered this genus as monotypic, after E. loncholoma was ascribed to Brassica subgen. Brassicaria (Gómez-Campo, 1999) and E. setulosa was moved to the proposed genus Guenthera (Gómez-Campo, 2003). Pratap & Gupta (2009) also supported this view, meanwhile Warwick, Francis & Gugel (2009) recognized four taxa: E. loncholoma, E. pinnatifida, E. setulosa and E. vesicaria. However, after E. pinnatifida was classified as a subspecies of E. vesicaria, http://www.theplantlist.org/ accepted only three species in this genus: E. loncholoma, E. setulosa and E. vesicaria, with this last species presenting three subspecies. Of these subspecies, two (E. vesicaria vesicaria and E. vesicaria pinnatifida) are circumscribed to the West Mediterranean region, whereas the subspecies sativa shows a circum-mediterranean distribution (Pignone & Gómez-Campo, 2011), although it is currently cultivated in many other areas of the world (Gómez-Campo & Prakash, 1999). Our phylogenetic analyses support the inclusion of M. foleyi in the genus Eruca (Figs. 1–4). In addition, other evidences separate M. foleyi from the genus Moricandia. This species has been reported to be 2n = 14 (Sobrino Vesperinas, 1997), clearly different from the caryotipic values reported for the rest of the Moricandia species (2n = 28, 56, 84; Sobrino Vesperinas, 1984), but also a different value when compared to the Eruca species (2n = 22; Harberd, 1976). Recently, in an ITS phylogeny, Schlüter et al. (2016) separated M. foleyi of the Moricandia lineage, although unfortunately they did not include in their analyses any Eruca samples. Anecdotally, Maire (1967) described M. foleyi as a glabrous and robust green herb with a strong smell of E. vesicaria (“à forte odeur d’Eruca vesicaria”). In our nuclear-sequences trees (24-samples and the GenBank-ITS sets) M. foleyi consistently appeared in the Eruca clade. The same pattern appeared in the cpDNA tree (see Fig. 2 and Fig. S2), indicating that both nuclear and cytoplasmic genes ascribe M. foleyi to the genus Eruca and excluding a recent hybridization event. Therefore, M. foleyi should be removed from the genus Moricandia and ascribed, at least provisionally, to the genus Eruca: Eruca foleyi (new combination): Eruca foleyi (Batt.) Lorite, Perfectti, Gómez, González-Megías & Abdelaziz comb. nov. urn:lsid:ipni.org:names:77166016-1 (see Appendix for a formal taxonomical description).
Conclusions
We have reported an inclusive dated phylogeny of the genus Moricandia, showing that, after the adscription of M. foleyi to the genus Eruca, it is a recent monophyletic genus that evolved in North Africa between 5.81 and 7.89 Ma diverging from its sister monotypic genus Rytidocarpus. Following the new aperture of the Strait of Gibraltar during the Zanclean period, Moricandia diverged in two different lineages: the Iberian and Afrian clades. At a finer scale, future work should address the evolutionary relationships between the different subspecies of M. arvensis and confirm the phylogenetic position of M. sinaica.
Supplemental Information
Supplementary information
Supplementary information contains:
Table S1: GenBank accessions.
Table S2: Comparison between the proposed new species Moricandia rytidocarpoides and Rytidocarpus moricandioides (N. Africa) for several quantitative morphological traits.
Table S3: Comparison of the proposed new species Moricandia rytidocarpoides with the most related species within the genus and with Rytidocarpus moricandioides from North Africa.
Table S4: Additional examined specimens.
Microsoft Word - Supplementary Material-Moricandia phylogeny-R01.docx
Figure S1: Maximum likelihood tree produced with the complete 24-samples set. Branch labels represent bootstrap percentage values. Scale: mean expected rates of substitution per site.
Figure S2: Tanglegram showing the maximum likelihood inference trees obtained from cpDNA (ndhF + trnT–trnF) and nuclear (ITS1 + ITS2) sequences.
Figure S3: Biplots showing the result of non-metric multidimensional scaling (NMDS) for individuals (left) and variables (right) performed with the morphological data presented in Supplementary table 2. Model data in the left are referred to the result of a Permutational Multivariate analysis using species as factor.
Figure S4: Microphotography of fruits of M. rytidocarpoides (left) and R. moricandioides (right). Fruits (A and B) and detail of fruit valve (C and D). Central insert (C) shows size differences of the fruit.
Sequence alignment for the 24-samples set
Sequence alignment, in nexus format, of the 24-samples set, including ITS1, ITS2, ndhF and trnT–trnF regions. It also includes the Moricandia sinaica ITS region.
Sequence alignment for the GenBank-ITS set
Sequence alignment (nexus format) for the ITS regions of the GenBank-ITS set.